Correlating SHAPE signatures with three-dimensional RNA structures
- PMID: 21752927
- PMCID: PMC3162334
- DOI: 10.1261/rna.2640111
Correlating SHAPE signatures with three-dimensional RNA structures
Abstract
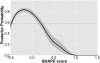
Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) is a facile technique for quantitative analysis of RNA secondary structure. In general, low SHAPE signal values indicate Watson-Crick base-pairing, and high values indicate positions that are single-stranded within the RNA structure. However, the relationship of SHAPE signals to structural properties such as non-Watson-Crick base-pairing or stacking has thus far not been thoroughly investigated. Here, we present results of SHAPE experiments performed on several RNAs with published three-dimensional structures. This strategy allows us to analyze the results in terms of correlations between chemical reactivities and structural properties of the respective nucleotide, such as different types of base-pairing, stacking, and phosphate-backbone interactions. We find that the RNA SHAPE signal is strongly correlated with cis-Watson-Crick/Watson-Crick base-pairing and is to a remarkable degree not dependent on other structural properties with the exception of stacking. We subsequently generated probabilistic models that estimate the likelihood that a residue with a given SHAPE score participates in base-pairing. We show that several models that take SHAPE scores of adjacent residues into account perform better in predicting base-pairing compared with individual SHAPE scores. This underscores the context sensitivity of SHAPE and provides a framework for an improved interpretation of the response of RNA to chemical modification.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
