A generic program for multistate protein design
- PMID: 21754981
- PMCID: PMC3130737
- DOI: 10.1371/journal.pone.0020937
A generic program for multistate protein design
Abstract
Some protein design tasks cannot be modeled by the traditional single state design strategy of finding a sequence that is optimal for a single fixed backbone. Such cases require multistate design, where a single sequence is threaded onto multiple backbones (states) and evaluated for its strengths and weaknesses on each backbone. For example, to design a protein that can switch between two specific conformations, it is necessary to to find a sequence that is compatible with both backbone conformations. We present in this paper a generic implementation of multistate design that is suited for a wide range of protein design tasks and demonstrate in silico its capabilities at two design tasks: one of redesigning an obligate homodimer into an obligate heterodimer such that the new monomers would not homodimerize, and one of redesigning a promiscuous interface to bind to only a single partner and to no longer bind the rest of its partners. Both tasks contained negative design in that multistate design was asked to find sequences that would produce high energies for several of the states being modeled. Success at negative design was assessed by computationally redocking the undesired protein-pair interactions; we found that multistate design's accuracy improved as the diversity of conformations for the undesired protein-pair interactions increased. The paper concludes with a discussion of the pitfalls of negative design, which has proven considerably more challenging than positive design.
Conflict of interest statement
Figures

 s for the homodimers vs the heterodimers comparing single state design (SSD) against multistate design (MSD). Binding energies were computed by redocking each of the complexes, and computing the difference between the lowest-energy from docking and the energies of the unbound monomers after their interface residues were allowed to pack. B) Histogram of the homodimer binding energy errors for each of the four rounds of multistate design. Errors were measured as the difference in the binding energies as computed by multistate design and as computed after redocking.
s for the homodimers vs the heterodimers comparing single state design (SSD) against multistate design (MSD). Binding energies were computed by redocking each of the complexes, and computing the difference between the lowest-energy from docking and the energies of the unbound monomers after their interface residues were allowed to pack. B) Histogram of the homodimer binding energy errors for each of the four rounds of multistate design. Errors were measured as the difference in the binding energies as computed by multistate design and as computed after redocking.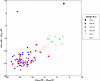

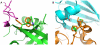
 atom on residue 279. The green rotamer collides with the chain E backbone (with an energy
atom on residue 279. The green rotamer collides with the chain E backbone (with an energy  5.1 REU) and, in the unbound, is pruned by Rosetta's bump check machinery (threshold of 5.0 REU); however, in the bound state, slightly favorable interactions with the chain I backbone rescue this rotamer by pushing its energy just beneath the bump-check threshold (
5.1 REU) and, in the unbound, is pruned by Rosetta's bump check machinery (threshold of 5.0 REU); however, in the bound state, slightly favorable interactions with the chain I backbone rescue this rotamer by pushing its energy just beneath the bump-check threshold ( 4.9 REU). Placing phenylalanine at 267 and anything besides glycine at 279 produces a large energy difference in the bound and unbound states which masquerades as an excellent binding energy.
4.9 REU). Placing phenylalanine at 267 and anything besides glycine at 279 produces a large energy difference in the bound and unbound states which masquerades as an excellent binding energy.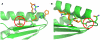
References
-
- Mandell D, Kortemme T. Computer-aided design of functional protein interactions. Nature Chemical Biology. 2009;5:797–807. - PubMed
-
- Dahiyat BI, Mayo SL. De Novo protein design: fully automated sequence selection. Science. 1997;278:82–87. - PubMed
-
- Malakauskas S, Mayo S. Design, structure and stability of a hyperthermophilic protein variant. Nature Structural Biology. 1998;5:470–475. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

