The repatterning of eukaryotic genomes by random genetic drift
- PMID: 21756106
- PMCID: PMC4519033
- DOI: 10.1146/annurev-genom-082410-101412
The repatterning of eukaryotic genomes by random genetic drift
Abstract
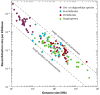
Recent observations on rates of mutation, recombination, and random genetic drift highlight the dramatic ways in which fundamental evolutionary processes vary across the divide between unicellular microbes and multicellular eukaryotes. Moreover, population-genetic theory suggests that the range of variation in these parameters is sufficient to explain the evolutionary diversification of many aspects of genome size and gene structure found among phylogenetic lineages. Most notably, large eukaryotic organisms that experience elevated magnitudes of random genetic drift are susceptible to the passive accumulation of mutationally hazardous DNA that would otherwise be eliminated by efficient selection. Substantial evidence also suggests that variation in the population-genetic environment influences patterns of protein evolution, with the emergence of certain kinds of amino-acid substitutions and protein-protein complexes only being possible in populations with relatively small effective sizes. These observations imply that the ultimate origins of many of the major genomic and proteomic disparities between prokaryotes and eukaryotes and among eukaryotic lineages have been molded as much by intrinsic variation in the genetic and cellular features of species as by external ecological forces.
Figures


References
-
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. - PubMed
-
- Bachtrog D. Expression profile of a degenerating neo-Y chromosome in Drosophila. Curr Biol. 2006;16:1694–99. - PubMed
-
- Baer CF, Miyamoto MM, Denver DR. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat Rev Genet. 2007;8:619–31. - PubMed
-
- Barraclough TG, Fontaneto D, Ricci C, Herniou EA. Evidence for inefficient selection against deleterious mutations in cytochrome oxidase I of asexual bdelloid rotifers. Mol Biol Evol. 2007;24:1952–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

