Phosphorylation of Serine 114 on Atg32 mediates mitophagy
- PMID: 21757540
- PMCID: PMC3164466
- DOI: 10.1091/mbc.E11-02-0145
Phosphorylation of Serine 114 on Atg32 mediates mitophagy
Abstract
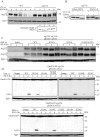
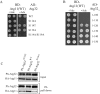
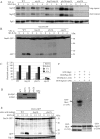
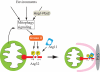
Mitophagy, which selectively degrades mitochondria via autophagy, has a significant role in mitochondrial quality control. When mitophagy is induced in yeast, mitochondrial residential protein Atg32 binds Atg11, an adaptor protein for selective types of autophagy, and it is recruited into the vacuole along with mitochondria. The Atg11-Atg32 interaction is believed to be the initial molecular step in which the autophagic machinery recognizes mitochondria as a cargo, although how this interaction is mediated is poorly understood. Therefore, we studied the Atg11-Atg32 interaction in detail. We found that the C-terminus region of Atg11, which included the fourth coiled-coil domain, interacted with the N-terminus region of Atg32 (residues 100-120). When mitophagy was induced, Ser-114 and Ser-119 on Atg32 were phosphorylated, and then the phosphorylation of Atg32, especially phosphorylation of Ser-114 on Atg32, mediated the Atg11-Atg32 interaction and mitophagy. These findings suggest that cells can regulate the amount of mitochondria, or select specific mitochondria (damaged or aged) that are degraded by mitophagy, by controlling the activity and/or localization of the kinase that phosphorylates Atg32. We also found that Hog1 and Pbs2, which are involved in the osmoregulatory signal transduction cascade, are related to Atg32 phosphorylation and mitophagy.
Figures


References
-
- Arai M, Imai H, Koumura T, Yoshida M, Emoto K, Umeda M, Chiba N, Nakagawa Y. Mitochondrial phospholipid hydroperoxide glutathione peroxidase plays a major role in preventing oxidative injury to cells. J Biol Chem. 1999;274:4924–4933. - PubMed
-
- Clotet J, Posas F. Control of cell cycle in response to osmostress: lessons from yeast. Methods Enzymol. 2007;428:63–76. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

