Identification of cis- and trans-acting genetic variants explaining up to half the variation in circulating vascular endothelial growth factor levels
- PMID: 21757650
- PMCID: PMC3193930
- DOI: 10.1161/CIRCRESAHA.111.243790
Identification of cis- and trans-acting genetic variants explaining up to half the variation in circulating vascular endothelial growth factor levels
Abstract
Rationale: Vascular endothelial growth factor (VEGF) affects angiogenesis, atherosclerosis, and cancer. Although the heritability of circulating VEGF levels is high, little is known about its genetic underpinnings.
Objective: Our aim was to identify genetic variants associated with circulating VEGF levels, using an unbiased genome-wide approach, and to explore their functional significance with gene expression and pathway analysis.
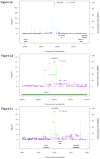
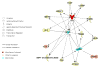
Methods and results: We undertook a genome-wide association study of serum VEGF levels in 3527 participants of the Framingham Heart Study, with preplanned replication in 1727 participants from 2 independent samples, the STANISLAS Family Study and the Prospective Investigation of the Vasculature in Uppsala Seniors study. One hundred forty single nucleotide polymorphism (SNPs) reached genome-wide significance (P<5×10(-8)). We found evidence of replication for the most significant associations in both replication datasets. In a conditional genome-wide association study, 4 SNPs mapping to 3 chromosomal regions were independently associated with circulating VEGF levels: rs6921438 and rs4416670 (6p21.1, P=6.11×10(-506) and P=1.47×10(-12)), rs6993770 (8q23.1, P=2.50×10(-16)), and rs10738760 (9p24.2, P=1.96×10(-34)). A genetic score including these 4 SNPs explained 48% of the heritability of serum VEGF levels. Six of the SNPs that reached genome-wide significance in the genome-wide association study were significantly associated with VEGF messenger RNA levels in peripheral blood mononuclear cells. Ingenuity pathway analyses showed found plausible biological links between VEGF and 2 novel genes in these loci (ZFPM2 and VLDLR).
Conclusions: Genetic variants explaining up to half the heritability of serum VEGF levels were identified. These new insights provide important clues to the pathways regulating circulating VEGF levels.
Figures
Similar articles
-
Association between SNPs of Circulating Vascular Endothelial Growth Factor Levels, Hypercholesterolemia and Metabolic Syndrome.Medicina (Kaunas). 2019 Aug 11;55(8):464. doi: 10.3390/medicina55080464. Medicina (Kaunas). 2019. PMID: 31405227 Free PMC article.
-
Six Novel Loci Associated with Circulating VEGF Levels Identified by a Meta-analysis of Genome-Wide Association Studies.PLoS Genet. 2016 Feb 24;12(2):e1005874. doi: 10.1371/journal.pgen.1005874. eCollection 2016 Feb. PLoS Genet. 2016. PMID: 26910538 Free PMC article.
-
What is the contribution of two genetic variants regulating VEGF levels to type 2 diabetes risk and to microvascular complications?PLoS One. 2013;8(2):e55921. doi: 10.1371/journal.pone.0055921. Epub 2013 Feb 6. PLoS One. 2013. PMID: 23405237 Free PMC article.
-
Vascular Endothelial Growth Factor and Ischemic Heart Disease Risk: A Mendelian Randomization Study.J Am Heart Assoc. 2017 Aug 1;6(8):e005619. doi: 10.1161/JAHA.117.005619. J Am Heart Assoc. 2017. PMID: 28765276 Free PMC article.
-
Vascular endothelial growth factor (VEGF)-related single nucleotide polymorphisms rs10738760 and rs6921438 are not associated with diabetic retinopathy (DR) in Slovenian patients with type 2 diabetes mellitus (T2DM).Bosn J Basic Med Sci. 2017 Nov 20;17(4):328-332. doi: 10.17305/bjbms.2017.2068. Bosn J Basic Med Sci. 2017. PMID: 29055125 Free PMC article.
Cited by
-
A common variant highly associated with plasma VEGFA levels also contributes to the variation of both LDL-C and HDL-C.J Lipid Res. 2013 Feb;54(2):535-41. doi: 10.1194/jlr.P030551. Epub 2012 Dec 2. J Lipid Res. 2013. PMID: 23204297 Free PMC article.
-
Genetic variation determines VEGF-A plasma levels in cancer patients.Sci Rep. 2018 Nov 5;8(1):16332. doi: 10.1038/s41598-018-34506-4. Sci Rep. 2018. PMID: 30397360 Free PMC article.
-
International Genome-Wide Association Study Consortium Identifies Novel Loci Associated With Blood Pressure in Children and Adolescents.Circ Cardiovasc Genet. 2016 Jun;9(3):266-278. doi: 10.1161/CIRCGENETICS.115.001190. Epub 2016 Mar 11. Circ Cardiovasc Genet. 2016. PMID: 26969751 Free PMC article.
-
Association between SNPs of Circulating Vascular Endothelial Growth Factor Levels, Hypercholesterolemia and Metabolic Syndrome.Medicina (Kaunas). 2019 Aug 11;55(8):464. doi: 10.3390/medicina55080464. Medicina (Kaunas). 2019. PMID: 31405227 Free PMC article.
-
Vascular endothelial growth factor (VEGF)-related polymorphisms rs10738760 and rs6921438 are not risk factors for proliferative diabetic retinopathy (PDR) in patients with type 2 diabetes mellitus (T2DM).Bosn J Basic Med Sci. 2019 Feb 12;19(1):94-100. doi: 10.17305/bjbms.2018.3519. Bosn J Basic Med Sci. 2019. PMID: 30579324 Free PMC article.
References
-
- Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–676. - PubMed
-
- Ferrara N, Davis-Smyth T. The biology of vascular endothelial growth factor. Endocr Rev. 1997;18:4–25. - PubMed
-
- Wei W, Jin H, Chen ZW, Zioncheck TF, Yim AP, He GW. Vascular endothelial growth factor-induced nitric oxide- and PGI2-dependent relaxation in human internal mammary arteries: a comparative study with KDR and Flt-1 selective mutants. J Cardiovasc Pharmacol. 2004;44:615–621. - PubMed
-
- Zachary I. Neuroprotective role of vascular endothelial growth factor: signalling mechanisms, biological function, and therapeutic potential. Neurosignals. 2005;14:207–221. - PubMed
-
- Hojo Y, Ikeda U, Zhu Y, Okada M, Ueno S, Arakawa H, Fujikawa H, Katsuki T, Shimada K. Expression of vascular endothelial growth factor in patients with acute myocardial infarction. J Am Coll Cardiol. 2000;35:968–973. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS017950/NS/NINDS NIH HHS/United States
- R01 AG016495/AG/NIA NIH HHS/United States
- R01 HL077477/HL/NHLBI NIH HHS/United States
- HL093029/HL/NHLBI NIH HHS/United States
- AG08122/AG/NIA NIH HHS/United States
- R01 AG008122/AG/NIA NIH HHS/United States
- AG031287/AG/NIA NIH HHS/United States
- R01 AG033040/AG/NIA NIH HHS/United States
- R01 HL093328/HL/NHLBI NIH HHS/United States
- HL-077477/HL/NHLBI NIH HHS/United States
- R01 AG031287/AG/NIA NIH HHS/United States
- AG033040/AG/NIA NIH HHS/United States
- N01-HC-25195/HC/NHLBI NIH HHS/United States
- P30 AG013846/AG/NIA NIH HHS/United States
- AG033193/AG/NIA NIH HHS/United States
- R01 HL093029/HL/NHLBI NIH HHS/United States
- NS17950/NS/NINDS NIH HHS/United States
- K24 HL004334/HL/NHLBI NIH HHS/United States
- R01 HL077447/HL/NHLBI NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- R01 AG033193/AG/NIA NIH HHS/United States
- P30AG013846/AG/NIA NIH HHS/United States
- HL-K24-04334/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

