A role for prolyl 3-hydroxylase 2 in post-translational modification of fibril-forming collagens
- PMID: 21757687
- PMCID: PMC3162427
- DOI: 10.1074/jbc.M111.267906
A role for prolyl 3-hydroxylase 2 in post-translational modification of fibril-forming collagens
Abstract
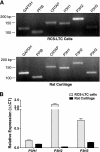
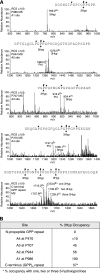
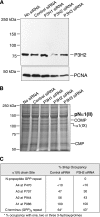
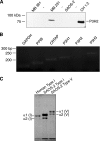
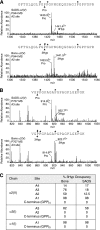
The fibrillar collagen types I, II, and V/XI have recently been shown to have partially 3-hydroxylated proline (3Hyp) residues at sites other than the established primary Pro-986 site in the collagen triple helical domain. These sites showed tissue specificity in degree of hydroxylation and a pattern of D-periodic spacing. This suggested a contributory role in fibril supramolecular assembly. The sites in clade A fibrillar α1(II), α2(V), and α1(I) collagen chains share common features with known prolyl 3-hydroxylase 2 (P3H2) substrate sites in α1(IV) chains implying a role for this enzyme. We pursued this possibility using the Swarm rat chondrosarcoma cell line (RCS-LTC) found to express high levels of P3H2 mRNA. Mass spectrometry determined that all the additional candidate 3Hyp substrate sites in the pN type II collagen made by these cells were highly hydroxylated. In RNA interference experiments, P3H2 protein synthesis was suppressed coordinately with prolyl 3-hydroxylation at Pro-944, Pro-707, and the C-terminal GPP repeat of the pNα1(II) chain, but Pro-986 remained fully hydroxylated. Furthermore, when P3H2 expression was turned off, as seen naturally in cultured SAOS-2 osteosarcoma cells, full 3Hyp occupancy at Pro-986 in α1(I) chains was unaffected, whereas 3-hydroxylation of residue Pro-944 in the α2(V) chain was largely lost, and 3-hydroxylation of Pro-707 in α2(V) and α2(I) chains were sharply reduced. The results imply that P3H2 has preferred substrate sequences among the classes of 3Hyp sites in clade A collagen chains.
Figures
References
-
- Olsen B. R., Berg R. A., Kivirikko K. T., Prockop D. J. (1973) Eur. J. Biochem. 35, 135–147 - PubMed
-
- Kivirikko K. I., Ryhänen L., Anttinen H., Bornstein P., Prockop D. J. (1973) Biochemistry 12, 4966–4971 - PubMed
-
- Vranka J. A., Sakai L. Y., Bächinger H. P. (2004) J. Biol. Chem. 279, 23615–23621 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

