The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates
- PMID: 21757688
- PMCID: PMC3162410
- DOI: 10.1074/jbc.M111.264721
The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates
Abstract
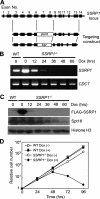
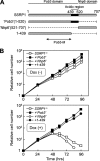
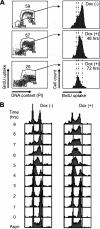
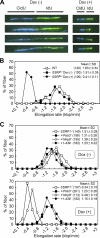
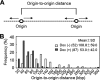
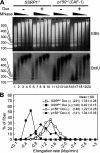
Ordered nucleosome disassembly and reassembly are required for eukaryotic DNA replication. The facilitates chromatin transcription (FACT) complex, a histone chaperone comprising Spt16 and SSRP1, is involved in DNA replication as well as transcription. FACT associates with the MCM helicase, which is involved in DNA replication initiation and elongation. Although the FACT-MCM complex is reported to regulate DNA replication initiation, its functional role in DNA replication elongation remains elusive. To elucidate the functional role of FACT in replication fork progression during DNA elongation in the cells, we generated and analyzed conditional SSRP1 gene knock-out chicken (Gallus gallus) DT40 cells. SSRP1-depleted cells ceased to grow and exhibited a delay in S-phase cell cycle progression, although SSRP1 depletion did not affect the level of chromatin-bound DNA polymerase α or nucleosome reassembly on daughter strands. The tracking length of newly synthesized DNA, but not origin firing, was reduced in SSRP1-depleted cells, suggesting that the S-phase cell cycle delay is mainly due to the inhibition of replication fork progression rather than to defects in the initiation of DNA replication in these cells. We discuss the mechanisms of how FACT promotes replication fork progression in the cells.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

