The T cell receptor triggering apparatus is composed of monovalent or monomeric proteins
- PMID: 21757710
- PMCID: PMC3173209
- DOI: 10.1074/jbc.M111.219212
The T cell receptor triggering apparatus is composed of monovalent or monomeric proteins
Abstract
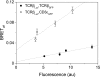
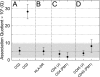
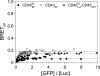
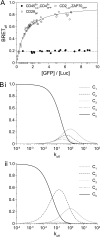
Understanding the component stoichiometry of the T cell antigen receptor (TCR) triggering apparatus is essential for building realistic models of signal initiation. Recent studies suggesting that the TCR and other signaling-associated proteins are preclustered on resting T cells relied on measurements of the behavior of membrane proteins at interfaces with functionalized glass surfaces. Using fluorescence recovery after photobleaching, we show that, compared with the apical surface, the mobility of TCRs is significantly reduced at Jurkat T cell/glass interfaces, in a signaling-sensitive manner. Using two biophysical approaches that mitigate these effects, bioluminescence resonance energy transfer and two-color coincidence detection microscopy, we show that, within the uncertainty of the methods, the membrane components of the TCR triggering apparatus, i.e. the TCR complex, MHC molecules, CD4/Lck and CD45, are exclusively monovalent or monomeric in human T cell lines, implying that TCR triggering depends only on the kinetics of TCR/pMHC interactions. These analyses also showed that constraining proteins to two dimensions at the cell surface greatly enhances random interactions versus those between the membrane and the cytoplasm. Simulations of TCR-pMHC complex formation based on these findings suggest how unclustered TCR triggering-associated proteins might nevertheless be capable of generating complex signaling outputs via the differential recruitment of cytosolic effectors to the cell membrane.
Figures
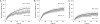

References
-
- Salzmann M., Bachmann M. F. (1998) Mol. Immunol. 35, 271–277 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

