Identification and characterization of a new true lipase isolated through metagenomic approach
- PMID: 21762508
- PMCID: PMC3161859
- DOI: 10.1186/1475-2859-10-54
Identification and characterization of a new true lipase isolated through metagenomic approach
Abstract
Background: Metagenomics, the application of molecular genomics to consortia of non-cultivated microbes, has the potential to have a substantial impact on the search for novel industrial enzymes such as esterases (carboxyl ester hydrolases, EC 3.1.1.1) and lipases (triacylglycerol lipases, EC 3.1.1.3). In the current work, a novel lipase gene was identified from a fosmid metagenomic library constructed with the "prokaryotic-enriched" DNA from a fat-contaminated soil collected from a wastewater treatment plant.
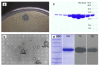
Results: In preliminary screening on agar containing 1% tributyrin, 2661 of the approximately 500,000 clones in the metagenomic library showed activity. Of these, 127 showed activity on agar containing 1% tricaprylin, while 32 were shown to be true lipase producers through screening on agar containing 1% triolein. The clone with the largest halo was further characterized. Its lipase gene showed 72% identity to a putative lipase of Yersinia enterocolitica subsp. palearctica Y11. The lipase, named LipC12, belongs to family I.1 of bacterial lipases, has a chaperone-independent folding, does not possess disulfide bridges and is calcium ion dependent. It is stable from pH 6 to 11 and has activity from pH 4.5 to 10, with higher activities at alkaline pH values. LipC12 is stable up to 3.7 M NaCl and from 20 to 50°C, with maximum activity at 30°C over a 1 h incubation. The pure enzyme has specific activities of 1722 U/mg and 1767 U/mg against olive oil and pig fat, respectively. Moreover, it is highly stable in organic solvents at 15% and 30% (v/v).
Conclusions: The combination of the use of a fat-contaminated soil, enrichment of prokaryotic DNA and a three-step screening strategy led to a high number of lipase-producing clones in the metagenomic library. The most notable properties of the new lipase that was isolated and characterized were a high specific activity against long chain triacylglycerols, activity and stability over a wide range of pH values, good thermal stability and stability in water-miscible organic solvents and at high salt concentrations. These characteristics suggest that this lipase has potential to perform well in biocatalytic processes, such as for hydrolysis and synthesis reactions involving long-chain triacylglycerols and fatty acid esters.
Figures


References
-
- Hasan F, Shah AA, Hameed A. Industrial applications of microbial lipases. Enzyme Microb Technol. 2006;39:235–251. doi: 10.1016/j.enzmictec.2005.10.016. - DOI
-
- Shu Z-Y, Jiang H, Lin R-F, Jiang Y-M, Lin L, Huang J-Z. Technical methods to improve yield, activity and stability in the development of microbial lipases. J Mol Catal B: Enzym. 2010;62:1–8. doi: 10.1016/j.molcatb.2009.09.003. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

