Comparative Proteomic Analysis of saccharopolyspora spinosa SP06081 and PR2 strains reveals the differentially expressed proteins correlated with the increase of spinosad yield
- PMID: 21762521
- PMCID: PMC3149565
- DOI: 10.1186/1477-5956-9-40
Comparative Proteomic Analysis of saccharopolyspora spinosa SP06081 and PR2 strains reveals the differentially expressed proteins correlated with the increase of spinosad yield
Abstract
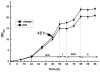
Background: Saccharopolyspora spinosa produces the environment-friendly biopesticide spinosad, a mixture of two polyketide-derived macrolide active ingredients called spinosyns A and D. Therefore considerable interest is in the improvement of spinosad production because of its low yield in wild-type S. spinosa. Recently, a spinosad-hyperproducing PR2 strain with stable heredity was obtained from protoplast regeneration of the wild-type S. spinosa SP06081 strain. A comparative proteomic analysis was performed on the two strains during the first rapid growth phase (RG1) in seed medium (SM) by using label-free quantitative proteomics to investigate the underlying mechanism leading to the enhancement of spinosad yield.
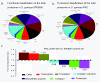
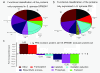
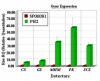
Results: In total, 224 proteins from the SP06081 strain and 204 proteins from the PR2 strain were unambiguously identified by liquid chromatography-tandem mass spectrometry analysis, sharing 140 proteins. A total of 12 proteins directly related to spinosad biosynthesis were identified from the two strains in RG1. Comparative analysis of the shared proteins revealed that approximately 31% of them changed their abundance significantly and fell in all of the functional groups, such as tricarboxylic acid cycles, glycolysis, biosynthetic processes, catabolic processes, transcription, translation, oxidation and reduction. Several key enzymes involved in the synthesis of primary metabolic intermediates used as precursors for spinosad production, energy supply, polyketide chain assembly, deoxysugar methylation, and antioxidative stress were differentially expressed in the same pattern of facilitating spinosad production by the PR2 strain. Real-time reverse transcriptase polymerase chain reaction analysis revealed that four of five selected genes showed a positive correlation between changes at the translational and transcriptional expression level, which further confirmed the proteomic analysis.
Conclusions: The present study is the first comprehensive and comparative proteome analysis of S. spinosa strains. Our results highlight the differentially expressed proteins between the two S. spinosa strains and provide some clues to understand the molecular and metabolic mechanisms that could lead to the increased spinosad production yield.
Figures


Similar articles
-
Differential proteomic profiling reveals regulatory proteins and novel links between primary metabolism and spinosad production in Saccharopolyspora spinosa.Microb Cell Fact. 2014 Feb 21;13(1):27. doi: 10.1186/1475-2859-13-27. Microb Cell Fact. 2014. PMID: 24555503 Free PMC article.
-
Effects of acuC on the growth development and spinosad biosynthesis of Saccharopolyspora spinosa.Microb Cell Fact. 2021 Jul 22;20(1):141. doi: 10.1186/s12934-021-01630-2. Microb Cell Fact. 2021. PMID: 34294095 Free PMC article.
-
Comparative transcriptomic analysis of two Saccharopolyspora spinosa strains reveals the relationships between primary metabolism and spinosad production.Sci Rep. 2021 Jul 20;11(1):14779. doi: 10.1038/s41598-021-94251-z. Sci Rep. 2021. PMID: 34285307 Free PMC article.
-
Strategies for Enhancing the Yield of the Potent Insecticide Spinosad in Actinomycetes.Biotechnol J. 2019 Jan;14(1):e1700769. doi: 10.1002/biot.201700769. Epub 2018 Jul 5. Biotechnol J. 2019. PMID: 29897659 Review.
-
Genes for the biosynthesis of spinosyns: applications for yield improvement in Saccharopolyspora spinosa.J Ind Microbiol Biotechnol. 2001 Dec;27(6):399-402. doi: 10.1038/sj.jim.7000180. J Ind Microbiol Biotechnol. 2001. PMID: 11774006 Review.
Cited by
-
High Level of Spinosad Production in the Heterologous Host Saccharopolyspora erythraea.Appl Environ Microbiol. 2016 Aug 30;82(18):5603-11. doi: 10.1128/AEM.00618-16. Print 2016 Sep 15. Appl Environ Microbiol. 2016. PMID: 27401975 Free PMC article.
-
Differential proteomic profiling reveals regulatory proteins and novel links between primary metabolism and spinosad production in Saccharopolyspora spinosa.Microb Cell Fact. 2014 Feb 21;13(1):27. doi: 10.1186/1475-2859-13-27. Microb Cell Fact. 2014. PMID: 24555503 Free PMC article.
-
Bacterioferritin: a key iron storage modulator that affects strain growth and butenyl-spinosyn biosynthesis in Saccharopolyspora pogona.Microb Cell Fact. 2021 Aug 14;20(1):157. doi: 10.1186/s12934-021-01651-x. Microb Cell Fact. 2021. PMID: 34391414 Free PMC article.
-
A High-Throughput Method Based on Microculture Technology for Screening of High-Yield Strains of Tylosin-Producing Streptomyces fradiae.J Microbiol Biotechnol. 2023 Jun 28;33(6):831-839. doi: 10.4014/jmb.2210.10023. Epub 2023 Mar 10. J Microbiol Biotechnol. 2023. PMID: 36994618 Free PMC article.
-
Effects of acuC on the growth development and spinosad biosynthesis of Saccharopolyspora spinosa.Microb Cell Fact. 2021 Jul 22;20(1):141. doi: 10.1186/s12934-021-01630-2. Microb Cell Fact. 2021. PMID: 34294095 Free PMC article.
References
-
- Kirst HA, Michel KH, Mynderse JS, Chio EH, Yao RC, Nakatsukasa WM, Boeck L, Occolowitz JL, Paschal JW, Deeter JB, Thompson GD. In: Synthesis and chemistry of agrochemicals. 1. Baker DR, Fenyes JG, Steffens JJ, editor. Vol. 3. Washington, DC: American Chemical Society; 1992. Discovery, isolation, and structure elucidation of a family of structurally unique fermentation derived tetracyclic macrolides; pp. 214–225.
-
- Kirst HA, Michel KH, Mynderse JS, Chio EH, Yao RC, Nakatsukasa WM, Boeck L, Occolowitz JL, Paschal JW, Deeter JB, Thompson GD. In: Development in industrial microbiology series: microbial metabolites. Brown WC, editor. Vol. 32. Washington, DC: Society for Industrial Microbiology; 1993. Discovery and identification of a novel fermentation derived insecticide; pp. 109–1161st edition.
LinkOut - more resources
Full Text Sources
Other Literature Sources

