Activation of the glutaredoxin-1 gene by nuclear factor κB enhances signaling
- PMID: 21762778
- PMCID: PMC3181077
- DOI: 10.1016/j.freeradbiomed.2011.06.025
Activation of the glutaredoxin-1 gene by nuclear factor κB enhances signaling
Abstract
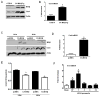
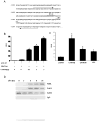
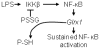
The transcription factor nuclear factor κB (NF-κB) is a critical regulator of inflammation and immunity and is negatively regulated via S-glutathionylation. The inhibitory effect of S-glutathionylation is overcome by glutaredoxin-1 (Grx1), which under physiological conditions catalyzes deglutathionylation and enhances NF-κB activation. The mechanisms whereby expression of the Glrx1 gene is regulated remain unknown. Here we examined the role of NF-κB in regulating activation of Glrx1. Transgenic mice that express a doxycycline-inducible constitutively active version of inhibitory κB kinase-β (CA-IKKβ) demonstrate elevated expression of Grx1. Transient transfection of CA-IKKβ also resulted in significant induction of Grx1. A 2-kb region of the Glrx1 promoter that contains two putative NF-κB binding sites was activated by CA-IKKβ, RelA/p50, and lipopolysaccharide (LPS). Chromatin immunoprecipitation experiments confirmed binding of RelA to the promoter of Glrx1 in response to LPS. Stimulation of C10 lung epithelial cells with LPS caused transient increases in Grx1 mRNA expression and time-dependent increases in S-glutathionylation of IKKβ. Overexpression of Grx1 decreased S-glutathionylation of IKKβ, prolonged NF-κB activation, and increased levels of proinflammatory mediators. Collectively, this study demonstrates that the Glrx1 gene is positively regulated by NF-κB and suggests a feed-forward mechanism to promote NF-κB signaling by decreasing S-glutathionylation.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




References
-
- Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–759. - PubMed
-
- Ghosh S, Hayden MS. New regulators of NF-kappaB in inflammation. Nat Rev Immunol. 2008;8:837–848. - PubMed
-
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. - PubMed
-
- Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3:221–227. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

