Novel cDNA sequences of aryl hydrocarbon receptors and gene expression in turtles (Chrysemys picta and Pseudemys scripta) exposed to different environments
- PMID: 21763458
- PMCID: PMC3176672
- DOI: 10.1016/j.cbpc.2011.06.017
Novel cDNA sequences of aryl hydrocarbon receptors and gene expression in turtles (Chrysemys picta and Pseudemys scripta) exposed to different environments
Abstract
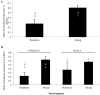
Reproductive changes have been observed in painted turtles from a site with known contamination located on Cape Cod, MA, USA. We hypothesize that these changes are caused by exposure to endocrine-disrupting compounds and that genes involved in reproduction are affected. The aryl hydrocarbon receptor (AHR) is an orphan receptor that is activated by environmental contaminants. AHR mRNA was measured in turtles exposed to soil collected from a contaminated site. Adult turtles were trapped from the study site (Moody Pond, MP) or a reference site and exposed to laboratory environments containing soil from either site. The red-eared slider was used to assess neonatal exposure to soil and water from the sites. The environmental exposures occurred over a 13-month period. Juveniles showed an age-dependent increase in brain AHR1. Juvenile turtles exposed to the MP environment had elevated gonadal AHR1. Adult turtles exposed to the MP environment showed significantly decreased brain AHR2. The painted turtle AHR is the first complete reptile AHR cDNA sequence. Phylogenetic analysis of the painted turtle AHR showed that it clusters with other AHR2s. Partial AHR1 and partial AHR2 cDNA sequences were cloned from the red-eared slider. MEME analysis identified 18 motifs in the turtle AHRs, showing high conservation between motifs that overlapped functional regions in both AHR isoforms.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures








References
-
- AFCEE. Massachusetts Military Reservation Plume Response Program, Prepared for AFCEE/MMR, Installation Restoration Program by Portage Ennvironmental Inc. Otis ANGB; MA, USA: 2003. US Air Force Center for Environmental Excellence (USAFCEE) Draft Final Second Five-year Review, 1998–2002 Massachusetts Military Reservation (MMR) Superfund Site, Otis Air National Guard Base, MA.
-
- Andreasen EA, Hahn ME, Heideman W, Peterson RE, Tanguay RL. The zebrafish (Danio rerio) aryl hydrocarbon receptor type 1 is a novel vertebrate receptor. Mol Pharmacol. 2002;62:234–249. - PubMed
-
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

