The conformation and orientation of a 27-residue CCR5 peptide in a ternary complex with HIV-1 gp120 and a CD4-mimic peptide
- PMID: 21763489
- PMCID: PMC3139148
- DOI: 10.1016/j.jmb.2011.04.023
The conformation and orientation of a 27-residue CCR5 peptide in a ternary complex with HIV-1 gp120 and a CD4-mimic peptide
Erratum in
- J Mol Biol. 2012 Apr 20;418(1-2):127
Abstract
Interaction of CC chemokine receptor 5 (CCR5) with the human immunodeficiency virus type 1 (HIV-1) gp120/CD4 complex involves its amino-terminal domain (Nt-CCR5) and requires sulfation of two to four tyrosine residues in Nt-CCR5. The conformation of a 27-residue Nt-CCR5 peptide, sulfated at Y10 and Y14, was studied both in its free form and in a ternary complex with deglycosylated gp120 and a CD4-mimic peptide. NMR experiments revealed a helical conformation at the center of Nt-CCR5(1-27), which is induced upon gp120 binding, as well as a helical propensity for the free peptide. A well-defined structure for the bound peptide was determined for residues 7-23, increasing by 2-fold the length of Nt-CCR5's known structure. Two-dimensional saturation transfer experiments and measurement of relaxation times highlighted Nt-CCR5 residues Y3, V5, P8-T16, E18, I23 and possibly D2 as the main binding determinant. A calculated docking model for Nt-CCR5(1-27) suggests that residues 2-22 of Nt-CCR5 interact with the bases of V3 and C4, while the C-terminal segment of Nt-CCR5(1-27) points toward the target cell membrane, reflecting an Nt-CCR5 orientation that differs by 180° from that of a previous model. A gp120 site that could accommodate (CCR5)Y3 in a sulfated form has been identified. The present model attributes a structural basis for binding interactions to all gp120 residues previously implicated in Nt-CCR5 binding. Moreover, the strong interaction of sulfated (CCR5)Tyr14 with (gp120)Arg440 revealed by the model and the previously found correlation between E322 and R440 mutations shed light on the role of these residues in HIV-1 phenotype conversion, furthering our understanding of CCR5 recognition by HIV-1.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures



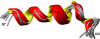



References
-
- Freed EO, Martin MA. The role of human immunodeficiency virus type 1 envelope glycoproteins in virus infection. J. Biol. Chem. 1995;270:23883–23886. - PubMed
-
- Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. - PubMed
-
- Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. - PubMed
-
- Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. - PubMed
-
- Chan DC, Kim PS. HIV entry and its inhibition. Cell. 1998;93:681–684. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

