T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway
- PMID: 21763495
- PMCID: PMC3139146
- DOI: 10.1016/j.jmb.2011.03.054
T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway
Abstract
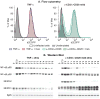
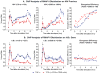
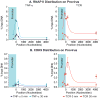
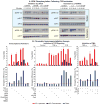
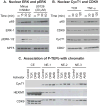
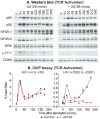
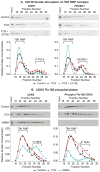
Latent human immunodeficiency virus (HIV) proviruses are thought to be primarily reactivated in vivo through stimulation of the T-cell receptor (TCR). Activation of the TCR induces multiple signal transduction pathways, leading to the ordered nuclear migration of the HIV transcription initiation factors NF-κB (nuclear factor κB) and NFAT (nuclear factor of activated T-cells), as well as potential effects on HIV transcriptional elongation. We have monitored the kinetics of proviral reactivation using chromatin immunoprecipitation assays to measure changes in the distribution of RNA polymerase II in the HIV provirus. Surprisingly, in contrast to TNF-α (tumor necrosis factor α) activation, where early transcription elongation is highly restricted due to rate-limiting concentrations of Tat, efficient and sustained HIV elongation and positive transcription elongation factor b (P-TEFb) recruitment are detected immediately after the activation of latent proviruses through the TCR. Inhibition of NFAT activation by cyclosporine had no effect on either HIV transcription initiation or elongation. However, examination of P-TEFb complexes by gel-filtration chromatography showed that TCR signaling led to the rapid dissociation of the large inactive P-TEFb:7SK RNP (small nuclear RNA 7SK ribonucleoprotein) complex and the release of active low-molecular-weight P-TEFb complexes. Both P-TEFb recruitment to the HIV long terminal repeat and enhanced HIV processivity were blocked by the ERK (extracellular-signal-regulated kinase) inhibitor U0126, but not by AKT (serine/threonine protein kinase Akt) and PI3K (phosphatidylinositol 3-kinase) inhibitors. In contrast to treatment with HMBA (hexamethylene bisacetamide) and DRB (5,6-dichlorobenzimidazole 1-β-ribofuranoside), which disrupt the large 7SK RNP complex but do not stimulate early HIV elongation, TCR signaling provides the first example of a physiological pathway that can shift the balance between the inactive P-TEFb pool and the active P-TEFb pool and thereby stimulate proviral reactivation.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures



References
-
- Pierson T, McArthur J, Siliciano RF. Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu Rev Immunol. 2000;18:665–708. - PubMed
-
- Coiras M, Lopez-Huertas MR, Perez-Olmeda M, Alcami J. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat Rev Microbiol. 2009;7:798–812. - PubMed
-
- Chun TW, Davey RT, Jr, Engel D, Lane HC, Fauci AS. Re-emergence of HIV after stopping therapy. Nature. 1999;401:874–5. - PubMed
-
- Davey RT, Jr, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, Kovacs JA, Polis MA, Walker RE, Falloon J, Masur H, Gee D, Baseler M, Dimitrov DS, Fauci AS, Lane HC. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci U S A. 1999;96:15109–14. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

