Resistance of bulky DNA lesions to nucleotide excision repair can result from extensive aromatic lesion-base stacking interactions
- PMID: 21764772
- PMCID: PMC3203604
- DOI: 10.1093/nar/gkr537
Resistance of bulky DNA lesions to nucleotide excision repair can result from extensive aromatic lesion-base stacking interactions
Abstract
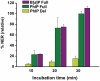
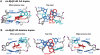
The molecular basis of resistance to nucleotide excision repair (NER) of certain bulky DNA lesions is poorly understood. To address this issue, we have studied NER in human HeLa cell extracts of two topologically distinct lesions, one derived from benzo[a]pyrene (10R-(+)-cis-anti-B[a]P-N(2)-dG), and one from the food mutagen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (C8-dG-PhIP), embedded in either full or 'deletion' duplexes (the partner nucleotide opposite the lesion is missing). All lesions adopt base-displaced intercalated conformations. Both full duplexes are thermodynamically destabilized and are excellent substrates of NER. However, the identical 10R-(+)-cis-anti-B[a]P-N(2)-dG adduct in the deletion duplex dramatically enhances the thermal stability of this duplex, and is completely resistant to NER. Molecular dynamics simulations show that B[a]P lesion-induced distortion/destabilization is compensated by stabilizing aromatic ring system-base stacking interactions. In the C8-dG-PhIP-deletion duplex, the smaller size of the aromatic ring system and the mobile phenyl ring are less stabilizing and yield moderate NER efficiency. Thus, a partner nucleotide opposite the lesion is not an absolute requirement for the successful initiation of NER. Our observations are consistent with the hypothesis that carcinogen-base stacking interactions, which contribute to the local DNA stability, can prevent the successful insertion of an XPC β-hairpin into the duplex and the normal recruitment of other downstream NER factors.
Figures





Similar articles
-
Resistance to Nucleotide Excision Repair of Bulky Guanine Adducts Opposite Abasic Sites in DNA Duplexes and Relationships between Structure and Function.PLoS One. 2015 Sep 4;10(9):e0137124. doi: 10.1371/journal.pone.0137124. eCollection 2015. PLoS One. 2015. PMID: 26340000 Free PMC article.
-
The DNA damage-sensing NER repair factor XPC-RAD23B does not recognize bulky DNA lesions with a missing nucleotide opposite the lesion.DNA Repair (Amst). 2020 Dec;96:102985. doi: 10.1016/j.dnarep.2020.102985. Epub 2020 Oct 1. DNA Repair (Amst). 2020. PMID: 33035795 Free PMC article.
-
Distant neighbor base sequence context effects in human nucleotide excision repair of a benzo[a]pyrene-derived DNA lesion.J Mol Biol. 2010 Jun 11;399(3):397-409. doi: 10.1016/j.jmb.2010.04.004. Epub 2010 Apr 22. J Mol Biol. 2010. PMID: 20399214 Free PMC article.
-
Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery.Biopolymers. 2002 Nov 5;65(3):202-10. doi: 10.1002/bip.10239. Biopolymers. 2002. PMID: 12228925 Review.
-
Repair-Resistant DNA Lesions.Chem Res Toxicol. 2017 Aug 21;30(8):1517-1548. doi: 10.1021/acs.chemrestox.7b00128. Epub 2017 Aug 10. Chem Res Toxicol. 2017. PMID: 28750166 Free PMC article. Review.
Cited by
-
Differences in the Access of Lesions to the Nucleotide Excision Repair Machinery in Nucleosomes.Biochemistry. 2015 Jul 14;54(27):4181-5. doi: 10.1021/acs.biochem.5b00564. Epub 2015 Jun 30. Biochemistry. 2015. PMID: 26091016 Free PMC article.
-
Inhibition of E. coli RecQ Helicase Activity by Structurally Distinct DNA Lesions: Structure-Function Relationships.Int J Mol Sci. 2022 Dec 9;23(24):15654. doi: 10.3390/ijms232415654. Int J Mol Sci. 2022. PMID: 36555294 Free PMC article.
-
Lesion Sensing during Initial Binding by Yeast XPC/Rad4: Toward Predicting Resistance to Nucleotide Excision Repair.Chem Res Toxicol. 2018 Nov 19;31(11):1260-1268. doi: 10.1021/acs.chemrestox.8b00231. Epub 2018 Oct 22. Chem Res Toxicol. 2018. PMID: 30284444 Free PMC article.
-
Molecular dynamics simulations reveal how H3K56 acetylation impacts nucleosome structure to promote DNA exposure for lesion sensing.DNA Repair (Amst). 2021 Nov;107:103201. doi: 10.1016/j.dnarep.2021.103201. Epub 2021 Aug 8. DNA Repair (Amst). 2021. PMID: 34399316 Free PMC article.
-
DNA adducts of the tobacco carcinogens 2-amino-9H-pyrido[2,3-b]indole and 4-aminobiphenyl are formed at environmental exposure levels and persist in human hepatocytes.Chem Res Toxicol. 2013 Sep 16;26(9):1367-77. doi: 10.1021/tx4002226. Epub 2013 Aug 16. Chem Res Toxicol. 2013. PMID: 23898916 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

