Vacuolar protein sorting protein 13A, TtVPS13A, localizes to the tetrahymena thermophila phagosome membrane and is required for efficient phagocytosis
- PMID: 21764909
- PMCID: PMC3187053
- DOI: 10.1128/EC.05089-11
Vacuolar protein sorting protein 13A, TtVPS13A, localizes to the tetrahymena thermophila phagosome membrane and is required for efficient phagocytosis
Abstract
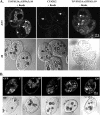
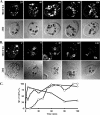
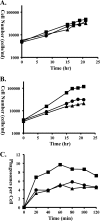
Vacuolar protein sorting 13 (VPS13) proteins have been studied in a number of organisms, and mutations in VPS13 genes have been implicated in two human genetic disorders, but the function of these proteins is poorly understood. The TtVPS13A protein was previously identified in a mass spectrometry analysis of the Tetrahymena thermophila phagosome proteome (M. E. Jacobs et al., Eukaryot. Cell 5:1990-2000, 2006), suggesting that it is involved in phagocytosis. In this study, we analyzed the structure of the macronuclear TtVPS13A gene, which was found to be composed of 17 exons spanning 12.5 kb and was predicted to encode a protein of 3,475 amino acids (aa). A strain expressing a TtVPS13A-green fluorescent protein (GFP) fusion protein was constructed, and the protein was found to associate with the phagosome membrane during the entire cycle of phagocytosis. In addition, Tetrahymena cells with a TtVPS13A knockout mutation displayed impaired phagocytosis. Specifically, they grew slowly under conditions where phagocytosis is essential, they formed few phagosomes, and the digestion of phagosomal contents was delayed compared to wild-type cells. Overall, these results provide evidence that the TtVPS13A protein is required for efficient phagocytosis.
Figures




Similar articles
-
The Tetrahymena thermophila phagosome proteome.Eukaryot Cell. 2006 Dec;5(12):1990-2000. doi: 10.1128/EC.00195-06. Epub 2006 Sep 29. Eukaryot Cell. 2006. PMID: 17012537 Free PMC article.
-
Directed motility of phagosomes in Tetrahymena thermophila requires actin and Myo1p, a novel unconventional myosin.Cell Motil Cytoskeleton. 2005 May;61(1):49-60. doi: 10.1002/cm.20065. Cell Motil Cytoskeleton. 2005. PMID: 15810016
-
Myo1 localizes to phagosomes, some of which traffic to the nucleus in a Myo1-dependent manner in Tetrahymena thermophila.Cell Motil Cytoskeleton. 2007 Dec;64(12):926-35. doi: 10.1002/cm.20233. Cell Motil Cytoskeleton. 2007. PMID: 17688250
-
Conservation and innovation in Tetrahymena membrane traffic: proteins, lipids, and compartments.Methods Cell Biol. 2012;109:141-75. doi: 10.1016/B978-0-12-385967-9.00006-2. Methods Cell Biol. 2012. PMID: 22444145 Free PMC article. Review.
-
Phagosome maturation: a few bugs in the system.J Membr Biol. 2003 Jun 1;193(3):137-52. doi: 10.1007/s00232-002-2008-2. J Membr Biol. 2003. PMID: 12962275 Review.
Cited by
-
VPS13 regulates membrane morphogenesis during sporulation in Saccharomyces cerevisiae.J Cell Sci. 2012 Jun 15;125(Pt 12):3004-11. doi: 10.1242/jcs.105114. Epub 2012 Mar 22. J Cell Sci. 2012. PMID: 22442115 Free PMC article.
-
Untangling the Thorns: Advances in the Neuroacanthocytosis Syndromes.J Mov Disord. 2015 May;8(2):41-54. doi: 10.14802/jmd.15009. Epub 2015 May 31. J Mov Disord. 2015. PMID: 26090076 Free PMC article. Review.
-
Autophagy in Dictyostelium: Mechanisms, regulation and disease in a simple biomedical model.Autophagy. 2017 Jan 2;13(1):24-40. doi: 10.1080/15548627.2016.1226737. Epub 2016 Oct 7. Autophagy. 2017. PMID: 27715405 Free PMC article. Review.
-
Human VPS13A is associated with multiple organelles and influences mitochondrial morphology and lipid droplet motility.Elife. 2019 Feb 11;8:e43561. doi: 10.7554/eLife.43561. Elife. 2019. PMID: 30741634 Free PMC article.
-
Eighth International Chorea-Acanthocytosis Symposium: Summary of Workshop Discussion and Action Points.Tremor Other Hyperkinet Mov (N Y). 2017 Feb 15;7:428. doi: 10.7916/D8XD127W. eCollection 2017. Tremor Other Hyperkinet Mov (N Y). 2017. PMID: 28224046 Free PMC article.
References
-
- Allen L. A., Aderem A. 1996. Mechanisms of phagocytosis. Curr. Opin. Immunol. 8:36–40 - PubMed
-
- Allen R. D., Fok A. K. 2000. Membrane trafficking and processing in Paramecium. Int. Rev. Cytol. 198:277–318 - PubMed
-
- Allen R. D., Wolf R. W. 1979. Membrane recycling at the cytoproct of Tetrahymena. J. Cell Sci. 35:217–227 - PubMed
-
- Bowen S., et al. 2007. The phagocytic capacity of neurons. Eur. J. Neurosci. 25:2947–2955 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

