Membrane proteases and aminoglycoside antibiotic resistance
- PMID: 21764915
- PMCID: PMC3165699
- DOI: 10.1128/JB.05133-11
Membrane proteases and aminoglycoside antibiotic resistance
Abstract
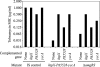
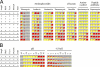
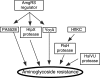
We present genetic studies that help define the functional network underlying intrinsic aminoglycoside resistance in Pseudomonas aeruginosa. Our analysis shows that proteolysis, particularly that controlled by the membrane protease FtsH, is a major determinant of resistance. First, we examined the consequences of inactivating genes controlled by AmgRS, a two-component regulator required for intrinsic tobramycin resistance. Three of the gene products account for resistance: a modulator of FtsH protease (YccA), a membrane protease (HtpX), and a membrane protein of unknown function (PA5528). Second, we screened mutations inactivating 66 predicted proteases and related functions. Insertions inactivating two FtsH protease accessory factors (HflK and HflC) and a cytoplasmic protease (HslUV) increased tobramycin sensitivity. Finally, we generated an ftsH deletion mutation. The mutation dramatically increased aminoglycoside sensitivity. Many of the functions whose inactivation increased sensitivity appeared to act independently, since multiple mutations led to additive or synergistic effects. Up to 500-fold increases in tobramycin sensitivity were observed. Most of the mutations also were highly pleiotropic, increasing sensitivity to a membrane protein hybrid, several classes of antibiotics, alkaline pH, NaCl, and other compounds. We propose that the network of proteases provides robust protection from aminoglycosides and other substances through the elimination of membrane-disruptive mistranslation products.
Copyright © 2011, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Akiyama Y. 2009. Quality control of cytoplasmic membrane proteins in Escherichia coli. J. Biochem. 146:449–454 - PubMed
-
- Bailey J., Manoil C. 2002. Genome-wide internal tagging of bacterial exported proteins. Nat. Biotechnol. 20:839–842 - PubMed
-
- Borovinskaya M. A., et al. 2007. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat. Struct. Mol. Biol. 14:727–732 - PubMed
-
- Choi K. H., et al. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2:443–448 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

