An integrated proteomics/transcriptomics approach points to oxygen as the main electron sink for methanol metabolism in Methylotenera mobilis
- PMID: 21764938
- PMCID: PMC3165657
- DOI: 10.1128/JB.05375-11
An integrated proteomics/transcriptomics approach points to oxygen as the main electron sink for methanol metabolism in Methylotenera mobilis
Abstract
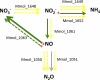
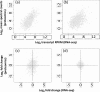
Methylotenera species, unlike their close relatives in the genera Methylophilus, Methylobacillus, and Methylovorus, neither exhibit the activity of methanol dehydrogenase nor possess mxaFI genes encoding this enzyme, yet they are able to grow on methanol. In this work, we integrated a genome-wide proteomics approach, shotgun proteomics, and a genome-wide transcriptomics approach, shotgun transcriptome sequencing (RNA-seq), of Methylotenera mobilis JLW8 to identify genes and enzymes potentially involved in methanol oxidation, with special attention to alternative nitrogen sources, to address the question of whether nitrate could play a role as an electron acceptor in place of oxygen. Both proteomics and transcriptomics identified a limited number of genes and enzymes specifically responding to methanol. This set includes genes involved in oxidative stress response systems, a number of oxidoreductases, including XoxF-type alcohol dehydrogenases, a type II secretion system, and proteins without a predicted function. Nitrate stimulated expression of some genes in assimilatory nitrate reduction and denitrification pathways, while ammonium downregulated some of the nitrogen metabolism genes. However, none of these genes appeared to respond to methanol, which suggests that oxygen may be the main electron sink during growth on methanol. This study identifies initial targets for future focused physiological studies, including mutant analysis, which will provide further details into this novel process.
Copyright © 2011, American Society for Microbiology. All Rights Reserved.
Figures

References
-
- Anthony C. 2004. The quinoprotein dehydrogenases for methanol and glucose. Arch. Biochem. Biophys. 428:2–9 - PubMed
-
- Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193 - PubMed
-
- Bosch G., et al. 2009. Insights into the physiology of Methylotenera mobilis as revealed by metagenome-based shotgun proteomic analysis. Microbiology 155:1103–1110 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

