Biodegradation of polyester polyurethane by endophytic fungi
- PMID: 21764951
- PMCID: PMC3165411
- DOI: 10.1128/AEM.00521-11
Biodegradation of polyester polyurethane by endophytic fungi
Abstract
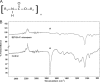
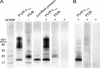
Bioremediation is an important approach to waste reduction that relies on biological processes to break down a variety of pollutants. This is made possible by the vast metabolic diversity of the microbial world. To explore this diversity for the breakdown of plastic, we screened several dozen endophytic fungi for their ability to degrade the synthetic polymer polyester polyurethane (PUR). Several organisms demonstrated the ability to efficiently degrade PUR in both solid and liquid suspensions. Particularly robust activity was observed among several isolates in the genus Pestalotiopsis, although it was not a universal feature of this genus. Two Pestalotiopsis microspora isolates were uniquely able to grow on PUR as the sole carbon source under both aerobic and anaerobic conditions. Molecular characterization of this activity suggests that a serine hydrolase is responsible for degradation of PUR. The broad distribution of activity observed and the unprecedented case of anaerobic growth using PUR as the sole carbon source suggest that endophytes are a promising source of biodiversity from which to screen for metabolic properties useful for bioremediation.
Figures





References
-
- Allen B. A., Hilliard N. P., Howard G. T. 1999. Purification and characterization of a soluble polyurethane degrading enzyme from Comamonas acidovorans. Int. Biodeterior. Biodegrad. 43:37–41
-
- Bacon C., White J. 2000. Microbial endophytes. Marcel Dekker, New York, NY
-
- Crabbe J. R., Campbell J. R., Thompson L., Walz S. L., Schultz W. W. 1994. Biodegradation of a colloidal ester-based polyurethane by soil fungi. Int. Biodeterior. Biodegrad. 33:103–113
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

