Cd-specific mutants of mercury-sensing regulatory protein MerR, generated by directed evolution
- PMID: 21764963
- PMCID: PMC3165431
- DOI: 10.1128/AEM.00662-11
Cd-specific mutants of mercury-sensing regulatory protein MerR, generated by directed evolution
Abstract
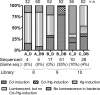
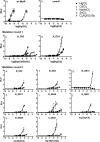
The mercury-sensing regulatory protein, MerR (Tn21), which regulates mercury resistance operons in Gram-negative bacteria, was subjected to directed evolution in an effort to generate a MerR mutant that responds to Cd but not Hg. Oligonucleotide-directed mutagenesis was used to introduce random mutations into the key metal-binding regions of MerR. The effects of these mutations were assessed using a vector in which MerR controlled the expression of green fluorescent protein (GFP) and luciferase via the mer operator/promoter. An Escherichia coli cell library was screened by fluorescence-activated cell sorting, using a fluorescence-based dual screening strategy that selected for MerR mutants that showed GFP repression when cells were induced with Hg but GFP activation in the presence of Cd. Two Cd-responsive MerR mutants with decreased responses toward Hg were identified through the first mutagenesis/selection round. These mutants were used for a second mutagenesis/selection round, which yielded eight Cd-specific mutants that had no significant response to Hg, Zn, or the other tested metal(loid)s. Seven of the eight Cd-specific MerR mutants showed repressor activities equal to that of wild-type (wt) MerR. These Cd-specific mutants harbored multiple mutations (12 to 22) in MerR, indicating that the alteration of metal specificity with maintenance of repressor function was due to the combined effect of many mutations rather than just a few amino acid changes. The amino acid changes were studied by alignment against the sequences of MerR and other metal-responsive MerR family proteins. The analysis indicated that the generated Cd-specific MerR mutants appear to be unique among the MerR family members characterized to date.
Figures


References
-
- Ansari A. Z., Chael M. L., O'Halloran T. V. 1992. Allosteric underwinding of DNA is a critical step in positive control of transcription by Hg-MerR. Nature 355:87–89 - PubMed
-
- Barrineau P., et al. 1984. The DNA sequence of the mercury resistance operon of the IncFII plasmid NR1. J. Mol. Appl. Genet. 2:601–619 - PubMed
-
- Brocklehurst K. R., et al. 1999. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol. Microbiol. 31:893–902 - PubMed
-
- Brocklehurst K. R., Megit S. J., Morby A. P. 2003. Characterisation of CadR from Pseudomonas aeruginosa: a Cd(II)-responsive MerR homologue. Biochem. Biophys. Res. Commun. 308:234–239 - PubMed
-
- Brown N. L., Ford S. J., Pridmore R. D., Fritzinger D. C. 1983. Nucleotide-sequence of a gene from the Pseudomonas transposon Tn501 encoding mercuric reductase. Biochemistry 22:4089–4095 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

