A limited microbial consortium is responsible for extended bioreduction of uranium in a contaminated aquifer
- PMID: 21764967
- PMCID: PMC3165427
- DOI: 10.1128/AEM.00220-11
A limited microbial consortium is responsible for extended bioreduction of uranium in a contaminated aquifer
Abstract
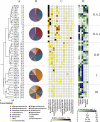
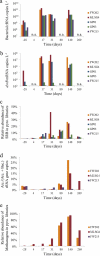
Subsurface amendments of slow-release substrates (e.g., emulsified vegetable oil [EVO]) are thought to be a pragmatic alternative to using short-lived, labile substrates for sustained uranium bioimmobilization within contaminated groundwater systems. Spatial and temporal dynamics of subsurface microbial communities during EVO amendment are unknown and likely differ significantly from those of populations stimulated by soluble substrates, such as ethanol and acetate. In this study, a one-time EVO injection resulted in decreased groundwater U concentrations that remained below initial levels for approximately 4 months. Pyrosequencing and quantitative PCR of 16S rRNA from monitoring well samples revealed a rapid decline in groundwater bacterial community richness and diversity after EVO injection, concurrent with increased 16S rRNA copy levels, indicating the selection of a narrow group of taxa rather than a broad community stimulation. Members of the Firmicutes family Veillonellaceae dominated after injection and most likely catalyzed the initial oil decomposition. Sulfate-reducing bacteria from the genus Desulforegula, known for long-chain fatty acid oxidation to acetate, also dominated after EVO amendment. Acetate and H(2) production during EVO degradation appeared to stimulate NO(3)(-), Fe(III), U(VI), and SO(4)(2-) reduction by members of the Comamonadaceae, Geobacteriaceae, and Desulfobacterales. Methanogenic archaea flourished late to comprise over 25% of the total microbial community. Bacterial diversity rebounded after 9 months, although community compositions remained distinct from the preamendment conditions. These results demonstrated that a one-time EVO amendment served as an effective electron donor source for in situ U(VI) bioreduction and that subsurface EVO degradation and metal reduction were likely mediated by successive identifiable guilds of organisms.
Figures





References
-
- Abdelouas A., Lutze W., Nuttall H. E. 1999. Uranium contamination in the subsurface; characterization and remediation. Rev. Mineral. Geochem. 38:433–473
-
- Akob D. M., Mills H. J., Kostka J. E. 2007. Metabolically active communities in uranium-contaminated subsurface sediments. FEMS Microbiol. Ecol. 59:95–107 - PubMed
-
- Atlas R. M., Horowitz A., Krichevsky M., Bej A. K. 1991. Response of microbial populations to environmental disturbance. Microb. Ecol. 22:249–256 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

