Novel virulent and broad-host-range Erwinia amylovora bacteriophages reveal a high degree of mosaicism and a relationship to Enterobacteriaceae phages
- PMID: 21764969
- PMCID: PMC3165370
- DOI: 10.1128/AEM.03022-10
Novel virulent and broad-host-range Erwinia amylovora bacteriophages reveal a high degree of mosaicism and a relationship to Enterobacteriaceae phages
Abstract
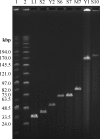
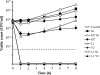
A diverse set of 24 novel phages infecting the fire blight pathogen Erwinia amylovora was isolated from fruit production environments in Switzerland. Based on initial screening, four phages (L1, M7, S6, and Y2) with broad host ranges were selected for detailed characterization and genome sequencing. Phage L1 is a member of the Podoviridae, with a 39.3-kbp genome featuring invariable genome ends with direct terminal repeats. Phage S6, another podovirus, was also found to possess direct terminal repeats but has a larger genome (74.7 kbp), and the virus particle exhibits a complex tail fiber structure. Phages M7 and Y2 both belong to the Myoviridae family and feature long, contractile tails and genomes of 84.7 kbp (M7) and 56.6 kbp (Y2), respectively, with direct terminal repeats. The architecture of all four phage genomes is typical for tailed phages, i.e., organized into function-specific gene clusters. All four phages completely lack genes or functions associated with lysogeny control, which correlates well with their broad host ranges and indicates strictly lytic (virulent) lifestyles without the possibility for host lysogenization. Comparative genomics revealed that M7 is similar to E. amylovora virus ΦEa21-4, whereas L1, S6, and Y2 are unrelated to any other E. amylovora phage. Instead, they feature similarities to enterobacterial viruses T7, N4, and ΦEcoM-GJ1. In a series of laboratory experiments, we provide proof of concept that specific two-phage cocktails offer the potential for biocontrol of the pathogen.
Figures


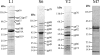


References
-
- Ackermann H.-W. 1996. Frequency of morphological phage descriptions in 1995. Arch. Virol. 141:209–218 - PubMed
-
- Adams M. H. 1959. Bacteriophages. Interscience Publishers Inc., New York, NY
-
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 - PubMed
-
- Balogh B., Canteros B. I., Stall R. E., Jones J. B. 2008. Control of citrus canker and citrus bacterial spot with bacteriophages. Plant Dis. 92:1048–1052 - PubMed
-
- Balogh B., Jones J. B., Iriarte F. B., Momol M. T. 2010. Phage therapy for plant disease control. Curr. Pharm. Biotechnol. 11:48–57 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

