Bayesian community-wide culture-independent microbial source tracking
- PMID: 21765408
- PMCID: PMC3791591
- DOI: 10.1038/nmeth.1650
Bayesian community-wide culture-independent microbial source tracking
Abstract
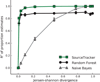
Contamination is a critical issue in high-throughput metagenomic studies, yet progress toward a comprehensive solution has been limited. We present SourceTracker, a Bayesian approach to estimate the proportion of contaminants in a given community that come from possible source environments. We applied SourceTracker to microbial surveys from neonatal intensive care units (NICUs), offices and molecular biology laboratories, and provide a database of known contaminants for future testing.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

