Dominant-negative mechanism of leukemogenic PAX5 fusions
- PMID: 21765475
- PMCID: PMC3197879
- DOI: 10.1038/onc.2011.291
Dominant-negative mechanism of leukemogenic PAX5 fusions
Abstract
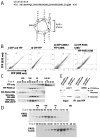
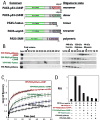
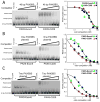
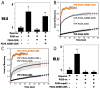
PAX5 encodes a master regulator of B-cell development. It fuses to other genes associated with acute lymphoblastoid leukemia (ALL). These fusion products are potent dominant-negative (DN) inhibitors of wild-type PAX5, resulting in a blockade of B-cell differentiation. Here, we show that multimerization of PAX5 DNA-binding domain (DBD) is necessary and sufficient to cause extremely stable chromatin binding and DN activity. ALL-associated PAX5-C20S results from fusion of the N-terminal region of PAX5, including its paired DBD, to the C-terminus of C20orf112, a protein of unknown function. We report that PAX5-C20S is a tetramer, which interacts extraordinarily stably with chromatin as determined by Fluorescence Recovery After Photobleaching in living cells. Tetramerization, stable chromatin binding and DN activity all require a putative five-turn amphipathic α-helix at the C-terminus of C20orf112, and does not require potential corepressor binding peptides elsewhere in the sequence. In vitro, the monomeric PAX5 DBD and PAX5-C20S binds a PAX5-binding site with equal affinity when it is at the center of an oligonucleotide too short to bind to more than one PAX5 DBD. But, PAX5-C20S binds the same sequence with 10-fold higher affinity than the monomeric PAX5 DBD when it is in a long DNA molecule. We suggest that the increased affinity results from interactions of one or more of the additional DBDs with neighboring non-specific sites in a long DNA molecule, and that this can account for the increased stability of PAX5-C20S chromatin binding compared with wild-type PAX5, resulting in DN activity by competition for binding to PAX5-target sites. Consistent with this model, the ALL-associated PAX5 fused to ETV6 or the multimerization domain of ETV6 SAM results in stable chromatin binding and DN activity. In addition, PAX5 DBD fused to artificial dimerization, trimerization and tetramerization domains results in parallel increases in the stability of chromatin binding and DN activity. Our studies suggest that oncogenic fusion proteins that retain the DBD of the transcription factor (TF) and the multimerization sequence of the partner protein can act in a DN manner by multimerizing and binding avidly to gene targets, preventing the normal TF from binding and inducing expression of its target genes. Inhibition of this multimeriztion may provide a novel therapeutic approach for cancers with this or similar fusion proteins.
Figures




Similar articles
-
Functional heterogeneity of PAX5 chimeras reveals insight for leukemia development.Mol Cancer Res. 2014 Apr;12(4):595-606. doi: 10.1158/1541-7786.MCR-13-0337. Epub 2014 Jan 16. Mol Cancer Res. 2014. PMID: 24435167
-
The reduced and altered activities of PAX5 are linked to the protein-protein interaction motif (coiled-coil domain) of the PAX5-PML fusion protein in t(9;15)-associated acute lymphocytic leukemia.Oncogene. 2011 Feb 24;30(8):967-77. doi: 10.1038/onc.2010.473. Epub 2010 Oct 25. Oncogene. 2011. PMID: 20972455
-
B Cell Linker Protein (BLNK) Is a Selective Target of Repression by PAX5-PML Protein in the Differentiation Block That Leads to the Development of Acute Lymphoblastic Leukemia.J Biol Chem. 2016 Feb 26;291(9):4723-31. doi: 10.1074/jbc.M115.637835. Epub 2015 Dec 24. J Biol Chem. 2016. PMID: 26703467 Free PMC article.
-
PAX5 fusion genes in acute lymphoblastic leukemia: A literature review.Medicine (Baltimore). 2023 May 19;102(20):e33836. doi: 10.1097/MD.0000000000033836. Medicine (Baltimore). 2023. PMID: 37335685 Free PMC article. Review.
-
Pax5: a master regulator of B cell development and leukemogenesis.Adv Immunol. 2011;111:179-206. doi: 10.1016/B978-0-12-385991-4.00005-2. Adv Immunol. 2011. PMID: 21970955 Review.
Cited by
-
PAX5-KIAA1549L: a novel fusion gene in a case of pediatric B-cell precursor acute lymphoblastic leukemia.Mol Cytogenet. 2015 Jul 8;8:48. doi: 10.1186/s13039-015-0138-3. eCollection 2015. Mol Cytogenet. 2015. PMID: 26157485 Free PMC article.
-
Mowat-Wilson Syndrome: Case Report and Review of ZEB2 Gene Variant Types, Protein Defects and Molecular Interactions.Int J Mol Sci. 2024 Feb 29;25(5):2838. doi: 10.3390/ijms25052838. Int J Mol Sci. 2024. PMID: 38474085 Free PMC article. Review.
-
Systematic Identification of Characteristic Genes of Ovarian Clear Cell Carcinoma Compared with High-Grade Serous Carcinoma Based on RNA-Sequencing.Int J Mol Sci. 2019 Sep 4;20(18):4330. doi: 10.3390/ijms20184330. Int J Mol Sci. 2019. PMID: 31487856 Free PMC article.
-
Activating PAX gene family paralogs to complement PAX5 leukemia driver mutations.PLoS Genet. 2018 Sep 14;14(9):e1007642. doi: 10.1371/journal.pgen.1007642. eCollection 2018 Sep. PLoS Genet. 2018. PMID: 30216339 Free PMC article.
-
Pax5 loss imposes a reversible differentiation block in B-progenitor acute lymphoblastic leukemia.Genes Dev. 2014 Jun 15;28(12):1337-50. doi: 10.1101/gad.240416.114. Genes Dev. 2014. PMID: 24939936 Free PMC article.
References
-
- Antolini F, Lo Bello M, Sette M. Purified promyelocytic leukemia coiled-coil aggregates as a tetramer displaying low alpha-helical content. Protein Expr Purif. 2003;29:94–102. - PubMed
-
- Bousquet M, Broccardo C, Quelen C, Meggetto F, Kuhlein E, Delsol G, et al. A novel PAX5-ELN fusion protein identified in B-cell acute lymphoblastic leukemia acts as a dominant negative on wild-type PAX5. Blood. 2007;109:3417–23. - PubMed
-
- Cazzaniga G, Daniotti M, Tosi S, Giudici G, Aloisi A, Pogliani E, et al. The paired box domain gene PAX5 is fused to ETV6/TEL in an acute lymphoblastic leukemia case. Cancer Res. 2001;61:4666–70. - PubMed
-
- Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

