Do all hemochromatosis patients have the same origin? An analysis of mitochondrial DNA and Y-DNA
- PMID: 21766093
- PMCID: PMC3142605
- DOI: 10.1155/2011/463810
Do all hemochromatosis patients have the same origin? An analysis of mitochondrial DNA and Y-DNA
Abstract
Background: Mitochondrial DNA (mtDNA) and Y-DNA analysis have been widely used to predict ancestral origin. Genetic anthropologists predict that human civilizations may have originated in central Africa one to two million years previously. Primary iron overload is not a common diagnosis among indigenous people of northern Africa, but hereditary hemochromatosis is present in approximately one in 200 people in northern Europe. MtDNA analysis has the potential to determine whether contemporary hemochromatosis patients have an ancient ancestral linkage.
Methods: DNA was obtained from buccal smears for mtDNA and Y-DNA analysis. Y-DNA analysis included examination of 20 short tandem repeat markers on the Y chromosome. Analysis of mtDNA involved sequencing of the HVR-1 genetic sequence (nucleotides 16001 to 16520) and was compared with the Cambridge Reference Sequence. MtDNA ancestral haplotypes were predicted from the analysis of the HVR-1 sequence.
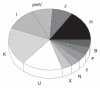
Results: Twenty-six male C282Y homozygotes were studied. There were 28 polymorphisms present in the HVR-1 sequence of these participants. The most common polymorphism was present at position 16519 in 15 participants and at position 16311 in eight participants. There were 12 different ancestral haplotypes predicted by mtDNA analysis, with the K haplotype being present in five participants. Y-DNA analysis revealed eight different haplotypes, with R1b being found in 11 of the 26 participants.
Conclusion: Analysis of mtDNA and Y-DNA in 26 hemochromatosis patients suggested that they did not all originate from the same ancestral tribe in Africa. These findings were consistent with the theory that the original hemochromatosis mutation occurred after migration of these ancestral people to central Europe, possibly 4000 years previously.
HISTORIQUE :: L’analyse de l’ADN mitochondrial (ADNmt) et de l’ADN-Y est largement utilisée pour prédire l’origine ancestrale. Selon les anthropologues génétiques, les civilisations humaines auraient vu le jour en Afrique centrale il y a de un à deux millions d’années. La surcharge en fer primaire n’est pas un diagnostic courant chez les peuples indigènes d’Afrique du Nord, mais on constate une hémochromatose héréditaire chez environ une personne sur 200 en Europe du Nord. L’analyse de l’ADNmt a le potentiel de déterminer si les patients actuels présentant une hémochromatose partagent la même origine ancestrale.
MÉTHODOLOGIE :: Les chercheurs ont fait un prélèvement buccal de l’ADN afin d’analyser l’ADNmt et l’ADN-Y des sujets. L’analyse de l’ADN-Y incluait l’examen de 20 marqueurs répétés courts sur le chromosome Y. L’analyse de l’ADNmt incluait le séquençage génétique HVR-1 (nucléotides 16001 à 16520) et a été comparée à la séquence de référence de Cambridge. Les chercheurs ont prédit les haplotypes d’ADNmt ancestral à partir de l’analyse de la séquence HVR-1.
RÉSULTATS :: Les chercheurs ont étudié 26 homozygotes C282Y mâles. Ils ont observé 28 polymorphismes dans la séquence HVR-1 de ces participants. Le principal polymorphisme était présent à la position 16519 chez 15 participants et à la position 16311 chez huit participants. Ils ont repéré 12 haplotypes ancestraux différents selon l’analyse de l’ADNmt, l’haplotype K étant présent chez cinq participants. L’analyse de l’ADN-Y a révélé huit haplotypes différents, le R1b étant présent chez 11 des 26 participants.
CONCLUSION :: L’analyse de l’ADNmt et de l’ADN-Y chez 26 patients ayant une hémochromatose indiquait qu’ils ne provenaient pas tous de la même tribu ancestrale d’Afrique. Ces observations correspondent à la théorie selon laquelle une mutation de l’hémochromatose originale s’est produite après la migration de ces peuples ancestraux en Europe centrale, il y a peut-être 4 000 ans.
Figures
Similar articles
-
Y-chromosome and mtDNA genetics reveal significant contrasts in affinities of modern Middle Eastern populations with European and African populations.PLoS One. 2013;8(1):e54616. doi: 10.1371/journal.pone.0054616. Epub 2013 Jan 30. PLoS One. 2013. PMID: 23382925 Free PMC article.
-
On the edge of Bantu expansions: mtDNA, Y chromosome and lactase persistence genetic variation in southwestern Angola.BMC Evol Biol. 2009 Apr 21;9:80. doi: 10.1186/1471-2148-9-80. BMC Evol Biol. 2009. PMID: 19383166 Free PMC article.
-
Larger mitochondrial DNA than Y-chromosome differences between matrilocal and patrilocal groups from Sumatra.Nat Commun. 2011;2:228. doi: 10.1038/ncomms1235. Nat Commun. 2011. PMID: 21407194
-
Out-of-Africa, the peopling of continents and islands: tracing uniparental gene trees across the map.Philos Trans R Soc Lond B Biol Sci. 2012 Mar 19;367(1590):770-84. doi: 10.1098/rstb.2011.0306. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22312044 Free PMC article. Review.
-
The archaeogenetics of Europe.Curr Biol. 2010 Feb 23;20(4):R174-83. doi: 10.1016/j.cub.2009.11.054. Curr Biol. 2010. PMID: 20178764 Review.
Cited by
-
Blood donor biobank as a resource in personalised biomedical genetic research.Eur J Hum Genet. 2024 Jan 12. doi: 10.1038/s41431-023-01528-0. Online ahead of print. Eur J Hum Genet. 2024. PMID: 38212662
-
HFE gene: Structure, function, mutations, and associated iron abnormalities.Gene. 2015 Dec 15;574(2):179-92. doi: 10.1016/j.gene.2015.10.009. Epub 2015 Oct 9. Gene. 2015. PMID: 26456104 Free PMC article. Review.
-
The evolutionary adaptation of the C282Y mutation to culture and climate during the European Neolithic.Am J Phys Anthropol. 2016 May;160(1):86-101. doi: 10.1002/ajpa.22937. Epub 2016 Jan 22. Am J Phys Anthropol. 2016. PMID: 26799452 Free PMC article.
References
-
- Adams PC, Barton JC. Haemochromatosis. Lancet. 2007;370:1855–60. - PubMed
-
- Adams PC, Reboussin DM, Barton JC, et al. Hemochromatosis and iron-overload screening in a racially diverse population. N Engl J Med. 2005;352:1769–78. - PubMed
-
- Whittington C. Was the C282Y mutation and Irish Gaelic mutation that the Vikings help disseminate? Medical Hypotheses. 2006;67:1270–3. - PubMed
-
- Lucotte G. Celtic origin of the C282Y mutation of hemochromatosis. Blood Cells Mol Dis. 1998;24:433–8. - PubMed
-
- Lucotte G, Dieterlen F. A European allele map of the C282Y mutation of hemochromatosis: Celtic versus Viking origin of the mutation? Blood Cells Mol Dis. 2003;31:262–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical