Direct quantification of the attempt frequency determining the mechanical unfolding of ubiquitin protein
- PMID: 21768096
- PMCID: PMC3173078
- DOI: 10.1074/jbc.M111.264093
Direct quantification of the attempt frequency determining the mechanical unfolding of ubiquitin protein
Abstract
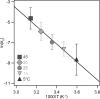
Understanding protein dynamics requires a comprehensive knowledge of the underlying potential energy surface that governs the motion of each individual protein molecule. Single molecule mechanical studies have provided the unprecedented opportunity to study the individual unfolding pathways along a well defined coordinate, the end-to-end length of the protein. In these experiments, unfolding requires surmounting an energy barrier that separates the native from the extended state. The calculation of the absolute value of the barrier height has traditionally relied on the assumption of an attempt frequency, υ(‡). Here we used single molecule force-clamp spectroscopy to directly determine the value of υ(‡) for mechanical unfolding by measuring the unfolding rate of the small protein ubiquitin at varying temperatures. Our experiments demonstrate a significant effect of the temperature on the mechanical rate of unfolding. By extrapolating the unfolding rate in the absence of force for different temperatures, varying within the range spanning from 5 to 45 °C, we measured a value for the activation barrier of ΔG(‡) = 71 ± 5 kJ/mol and an exponential prefactor υ(‡) ∼4 × 10(9) s(-1). Although the measured prefactor value is 3 orders of magnitude smaller than the value predicted by the transition state theory (∼6 × 10(12) s(-1)), it is 400-fold higher than that encountered in analogous experiments studying the effect of temperature on the reactivity of a protein-embedded disulfide bond (∼10(7) M(-1) s(-1)). This approach will allow quantitative characterization of the complete energy landscape of a folding polypeptide from highly extended states, of capital importance for proteins with elastic function.
Figures




References
-
- Jackson S. E. (1998) Fold. Des. 3, R81–R91 - PubMed
-
- Fersht A. (1999) Structure and Mechanism in Protein Science, W. H. Freeman, New York
-
- Eyring H. (1935) J. Chem. Phys. 3, 107–115
-
- Garcia-Viloca M., Gao J., Karplus M., Truhlar D. G. (2004) Science 303, 186–195 - PubMed
-
- Collet O., Chipot C. (2003) J. Am. Chem. Soc. 125, 6573–6580 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

