Lectin domains of polypeptide GalNAc transferases exhibit glycopeptide binding specificity
- PMID: 21768105
- PMCID: PMC3173194
- DOI: 10.1074/jbc.M111.273722
Lectin domains of polypeptide GalNAc transferases exhibit glycopeptide binding specificity
Abstract
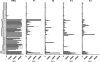
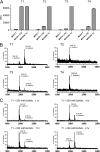
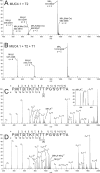
UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferases (GalNAc-Ts) constitute a family of up to 20 transferases that initiate mucin-type O-glycosylation. The transferases are structurally composed of catalytic and lectin domains. Two modes have been identified for the selection of glycosylation sites by GalNAc-Ts: confined sequence recognition by the catalytic domain alone, and concerted recognition of acceptor sites and adjacent GalNAc-glycosylated sites by the catalytic and lectin domains, respectively. Thus far, only the catalytic domain has been shown to have peptide sequence specificity, whereas the primary function of the lectin domain is to increase affinity to previously glycosylated substrates. Whether the lectin domain also has peptide sequence selectivity has remained unclear. Using a glycopeptide array with a library of synthetic and recombinant glycopeptides based on sequences of mucins MUC1, MUC2, MUC4, MUC5AC, MUC6, and MUC7 as well as a random glycopeptide bead library, we examined the binding properties of four different lectin domains. The lectin domains of GalNAc-T1, -T2, -T3, and -T4 bound different subsets of small glycopeptides. These results indicate an additional level of complexity in the initiation step of O-glycosylation by GalNAc-Ts.
Figures



Similar articles
-
Molecular Recognition of GalNAc in Mucin-Type O-Glycosylation.Acc Chem Res. 2023 Mar 7;56(5):548-560. doi: 10.1021/acs.accounts.2c00723. Epub 2023 Feb 23. Acc Chem Res. 2023. PMID: 36815693 Free PMC article. Review.
-
The lectin domain of the polypeptide GalNAc transferase family of glycosyltransferases (ppGalNAc Ts) acts as a switch directing glycopeptide substrate glycosylation in an N- or C-terminal direction, further controlling mucin type O-glycosylation.J Biol Chem. 2013 Jul 5;288(27):19900-14. doi: 10.1074/jbc.M113.477877. Epub 2013 May 20. J Biol Chem. 2013. PMID: 23689369 Free PMC article.
-
Mucin-type O-glycosylation is controlled by short- and long-range glycopeptide substrate recognition that varies among members of the polypeptide GalNAc transferase family.Glycobiology. 2016 Apr;26(4):360-76. doi: 10.1093/glycob/cwv108. Epub 2015 Nov 26. Glycobiology. 2016. PMID: 26610890 Free PMC article.
-
The catalytic and lectin domains of UDP-GalNAc:polypeptide alpha-N-Acetylgalactosaminyltransferase function in concert to direct glycosylation site selection.J Biol Chem. 2008 Aug 22;283(34):22942-51. doi: 10.1074/jbc.M803387200. Epub 2008 Jun 18. J Biol Chem. 2008. PMID: 18562306 Free PMC article.
-
Polypeptide GalNAc-Ts: from redundancy to specificity.Curr Opin Struct Biol. 2019 Jun;56:87-96. doi: 10.1016/j.sbi.2018.12.007. Epub 2019 Jan 28. Curr Opin Struct Biol. 2019. PMID: 30703750 Free PMC article. Review.
Cited by
-
A glycogene mutation map for discovery of diseases of glycosylation.Glycobiology. 2015 Feb;25(2):211-24. doi: 10.1093/glycob/cwu104. Epub 2014 Sep 28. Glycobiology. 2015. PMID: 25267602 Free PMC article.
-
Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family.Glycobiology. 2012 Jun;22(6):736-56. doi: 10.1093/glycob/cwr182. Epub 2011 Dec 18. Glycobiology. 2012. PMID: 22183981 Free PMC article. Review.
-
Molecular Recognition of GalNAc in Mucin-Type O-Glycosylation.Acc Chem Res. 2023 Mar 7;56(5):548-560. doi: 10.1021/acs.accounts.2c00723. Epub 2023 Feb 23. Acc Chem Res. 2023. PMID: 36815693 Free PMC article. Review.
-
Dynamic interplay between catalytic and lectin domains of GalNAc-transferases modulates protein O-glycosylation.Nat Commun. 2015 May 5;6:6937. doi: 10.1038/ncomms7937. Nat Commun. 2015. PMID: 25939779 Free PMC article.
-
Initiation of GalNAc-type O-glycosylation in the endoplasmic reticulum promotes cancer cell invasiveness.Proc Natl Acad Sci U S A. 2013 Aug 20;110(34):E3152-61. doi: 10.1073/pnas.1305269110. Epub 2013 Aug 2. Proc Natl Acad Sci U S A. 2013. PMID: 23912186 Free PMC article.
References
-
- Clausen H., Bennett E. P. (1996) Glycobiology 6, 635–646 - PubMed
-
- Wandall H. H., Hassan H., Mirgorodskaya E., Kristensen A. K., Roepstorff P., Bennett E. P., Nielsen P. A., Hollingsworth M. A., Burchell J., Taylor-Papadimitriou J., Clausen H. (1997) J. Biol. Chem. 272, 23503–23514 - PubMed
-
- Wojczyk B. S., Stwora-Wojczyk M. M., Hagen F. K., Striepen B., Hang H. C., Bertozzi C. R., Roos D. S., Spitalnik S. L. (2003) Mol. Biochem. Parasitol. 131, 93–107 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

