Unequal allelic expression of wild-type and mutated β-myosin in familial hypertrophic cardiomyopathy
- PMID: 21769673
- PMCID: PMC3228959
- DOI: 10.1007/s00395-011-0205-9
Unequal allelic expression of wild-type and mutated β-myosin in familial hypertrophic cardiomyopathy
Abstract
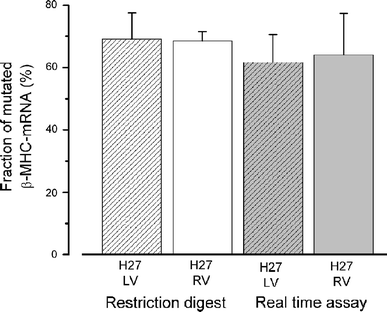
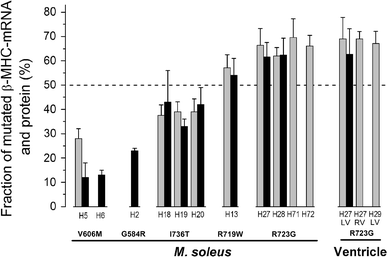
Familial hypertrophic cardiomyopathy (FHC) is an autosomal dominant disease, which in about 30% of the patients is caused by missense mutations in one allele of the β-myosin heavy chain (β-MHC) gene (MYH7). To address potential molecular mechanisms underlying the family-specific prognosis, we determined the relative expression of mutant versus wild-type MYH7-mRNA. We found a hitherto unknown mutation-dependent unequal expression of mutant to wild-type MYH7-mRNA, which is paralleled by similar unequal expression of β-MHC at the protein level. Relative abundance of mutated versus wild-type MYH7-mRNA was determined by a specific restriction digest approach and by real-time PCR (RT-qPCR). Fourteen samples from M. soleus and myocardium of 12 genotyped and clinically well-characterized FHC patients were analyzed. The fraction of mutated MYH7-mRNA in five patients with mutation R723G averaged to 66 and 68% of total MYH7-mRNA in soleus and myocardium, respectively. For mutations I736T, R719W and V606M, fractions of mutated MYH7-mRNA in M. soleus were 39, 57 and 29%, respectively. For all mutations, unequal abundance was similar at the protein level. Importantly, fractions of mutated transcripts were comparable among siblings, in younger relatives and unrelated carriers of the same mutation. Hence, the extent of unequal expression of mutated versus wild-type transcript and protein is characteristic for each mutation, implying cis-acting regulatory mechanisms. Bioinformatics suggest mRNA stability or splicing effectors to be affected by certain mutations. Intriguingly, we observed a correlation between disease expression and fraction of mutated mRNA and protein. This strongly suggests that mutation-specific allelic imbalance represents a new pathogenic factor for FHC.
Figures


Similar articles
-
Outcome of clinical versus genetic family screening in hypertrophic cardiomyopathy with focus on cardiac beta-myosin gene mutations.Cardiovasc Res. 2003 Feb;57(2):347-57. doi: 10.1016/s0008-6363(02)00711-3. Cardiovasc Res. 2003. PMID: 12566107
-
Intrinsic MYH7 expression regulation contributes to tissue level allelic imbalance in hypertrophic cardiomyopathy.J Muscle Res Cell Motil. 2017 Aug;38(3-4):291-302. doi: 10.1007/s10974-017-9486-4. Epub 2017 Nov 3. J Muscle Res Cell Motil. 2017. PMID: 29101517 Free PMC article.
-
The left and right ventricle of a patient with a R723G mutation of the beta-myosin heavy chain and severe hypertrophic cardiomyopathy show no differences in the expression of myosin mRNA.Cardiol J. 2010;17(5):518-22. Cardiol J. 2010. PMID: 20865685
-
Hypertrophic cardiomyopathy MYH7 mutation R723G alters mRNA secondary structure.Physiol Genomics. 2020 Jan 1;52(1):15-19. doi: 10.1152/physiolgenomics.00100.2019. Epub 2019 Dec 2. Physiol Genomics. 2020. PMID: 31790337
-
Altered force generation and cell-to-cell contractile imbalance in hypertrophic cardiomyopathy.Pflugers Arch. 2019 May;471(5):719-733. doi: 10.1007/s00424-019-02260-9. Epub 2019 Feb 11. Pflugers Arch. 2019. PMID: 30740621 Free PMC article. Review.
Cited by
-
Mechanical and kinetic properties of β-cardiac/slow skeletal muscle myosin.J Muscle Res Cell Motil. 2012 Dec;33(6):403-17. doi: 10.1007/s10974-012-9315-8. Epub 2012 Jul 31. J Muscle Res Cell Motil. 2012. PMID: 22847802
-
Two strikes and you're out: gene-gene mutation interactions in HCM.Circ Res. 2014 Jul 7;115(2):208-10. doi: 10.1161/CIRCRESAHA.114.304383. Circ Res. 2014. PMID: 24989488 Free PMC article. No abstract available.
-
Hypothesis and theory: mechanical instabilities and non-uniformities in hereditary sarcomere myopathies.Front Physiol. 2014 Sep 15;5:350. doi: 10.3389/fphys.2014.00350. eCollection 2014. Front Physiol. 2014. PMID: 25309450 Free PMC article.
-
Cardiomyopathies and Related Changes in Contractility of Human Heart Muscle.Int J Mol Sci. 2018 Jul 31;19(8):2234. doi: 10.3390/ijms19082234. Int J Mol Sci. 2018. PMID: 30065175 Free PMC article. Review.
-
Critical Evaluation of Current Hypotheses for the Pathogenesis of Hypertrophic Cardiomyopathy.Int J Mol Sci. 2022 Feb 16;23(4):2195. doi: 10.3390/ijms23042195. Int J Mol Sci. 2022. PMID: 35216312 Free PMC article. Review.
References
-
- Alcalai R, Seidman JG, Seidman CE. Genetic basis of hypertrophic cardiomyopathy: from bench to the clinics. J Cardiovasc Electrophysiol. 2008;19(1):104–110. - PubMed
-
- Arad M, Seidman JG, Seidman CE. Phenotypic diversity in hypertrophic cardiomyopathy. Hum Mol Genet. 2002;11(20):2499–2506. - PubMed
-
- Becker E, Navarro-Lopez F, Francino A, Brenner B, Kraft T. Quantification of mutant versus wild-type myosin in human muscle biopsies using nano-LC/ESI-MS. Anal Chem. 2007;79(24):9531–9538. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

