Cell-based quantification of molecular biomarkers in histopathology specimens
- PMID: 21771025
- PMCID: PMC3142095
- DOI: 10.1111/j.1365-2559.2011.03878.x
Cell-based quantification of molecular biomarkers in histopathology specimens
Abstract
Aims: To investigate the use of a computer-assisted technology for objective, cell-based quantification of molecular biomarkers in specified cell types in histopathology specimens, with the aim of advancing current visual estimation and pixel-level (rather than cell-based) quantification methods.
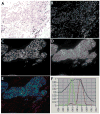
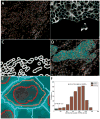
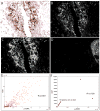
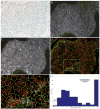
Methods and results: Tissue specimens were multiplex-immunostained to reveal cell structures, cell type markers, and analytes, and imaged with multispectral microscopy. The image data were processed with novel software that automatically delineates and types each cell in the field, measures morphological features, and quantifies analytes in different subcellular compartments of specified cells.The methodology was validated with the use of cell blocks composed of differentially labelled cultured cells mixed in known proportions, and evaluated on human breast carcinoma specimens for quantifying human epidermal growth factor receptor 2, estrogen receptor, progesterone receptor, Ki67, phospho-extracellular signal-related kinase, and phospho-S6. Automated cell-level analyses closely matched human assessments, but, predictably, differed from pixel-level analyses of the same images.
Conclusions: Our method reveals the type, distribution, morphology and biomarker state of each cell in the field, and allows multiple biomarkers to be quantified over specified cell types, regardless of their abundance. It is ideal for studying specimens from patients in clinical trials of targeted therapeutic agents, for investigating minority stromal cell subpopulations, and for phenotypic characterization to personalize therapy and prognosis.
© 2011 Blackwell Publishing Limited.
Figures


References
-
- Hammerschmied CG, Walter B, Hartmann A. Renal cell carcinoma 2008: histopathology, molecular genetics and new therapeutic options. Pathologe. 2008;29:354–363. - PubMed
-
- Crocker J, Murray P. Molecular biology in cellular pathology. Chichester: Wiley; 2003.
-
- Research Signpost (Trivandrum India) Recent research developments in histopathology. Trivandrum, India: Research Signpost; 2002. v.
-
- Menard S, Balsari A, Tagliabue E, et al. Biology, prognosis and response to therapy of breast carcinomas according to HER2 score. Ann Oncol. 2008;19:1706–1712. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

