Characterization of the HIV-1 RNA associated proteome identifies Matrin 3 as a nuclear cofactor of Rev function
- PMID: 21771346
- PMCID: PMC3160904
- DOI: 10.1186/1742-4690-8-60
Characterization of the HIV-1 RNA associated proteome identifies Matrin 3 as a nuclear cofactor of Rev function
Abstract
Background: Central to the fully competent replication cycle of the human immunodeficiency virus type 1 (HIV-1) is the nuclear export of unspliced and partially spliced RNAs mediated by the Rev posttranscriptional activator and the Rev response element (RRE).
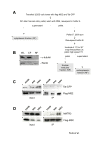
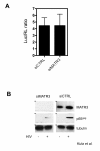
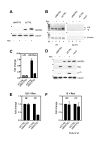
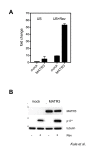
Results: Here, we introduce a novel method to explore the proteome associated with the nuclear HIV-1 RNAs. At the core of the method is the generation of cell lines harboring an integrated provirus carrying RNA binding sites for the MS2 bacteriophage protein. Flag-tagged MS2 is then used for affinity purification of the viral RNA. By this approach we found that the viral RNA is associated with the host nuclear matrix component MATR3 (Matrin 3) and that its modulation affected Rev activity. Knockdown of MATR3 suppressed Rev/RRE function in the export of unspliced HIV-1 RNAs. However, MATR3 was able to associate with Rev only through the presence of RRE-containing viral RNA.
Conclusions: In this work, we exploited a novel proteomic method to identify MATR3 as a cellular cofactor of Rev activity. MATR3 binds viral RNA and is required for the Rev/RRE mediated nuclear export of unspliced HIV-1 RNAs.
Figures

Similar articles
-
Matrin 3 is a co-factor for HIV-1 Rev in regulating post-transcriptional viral gene expression.Retrovirology. 2011 Jul 20;8:61. doi: 10.1186/1742-4690-8-61. Retrovirology. 2011. PMID: 21771347 Free PMC article.
-
HIV-1 pre-mRNA commitment to Rev mediated export through PSF and Matrin 3.Virology. 2013 Jan 20;435(2):329-40. doi: 10.1016/j.virol.2012.10.032. Epub 2012 Nov 13. Virology. 2013. PMID: 23158102
-
Posttranscriptional Regulation of HIV-1 Gene Expression during Replication and Reactivation from Latency by Nuclear Matrix Protein MATR3.mBio. 2018 Nov 13;9(6):e02158-18. doi: 10.1128/mBio.02158-18. mBio. 2018. PMID: 30425153 Free PMC article.
-
HIV Rev Assembly on the Rev Response Element (RRE): A Structural Perspective.Viruses. 2015 Jun 12;7(6):3053-75. doi: 10.3390/v7062760. Viruses. 2015. PMID: 26075509 Free PMC article. Review.
-
[HIV-1 Rev and related inhibitors].Yao Xue Xue Bao. 2007 Apr;42(4):347-51. Yao Xue Xue Bao. 2007. PMID: 17633198 Review. Chinese.
Cited by
-
Epigenetic silencing of Oct4 by a complex containing SUV39H1 and Oct4 pseudogene lncRNA.Nat Commun. 2015 Jul 9;6:7631. doi: 10.1038/ncomms8631. Nat Commun. 2015. PMID: 26158551 Free PMC article.
-
Matrin 3 is a co-factor for HIV-1 Rev in regulating post-transcriptional viral gene expression.Retrovirology. 2011 Jul 20;8:61. doi: 10.1186/1742-4690-8-61. Retrovirology. 2011. PMID: 21771347 Free PMC article.
-
Host restriction factors in retroviral infection: promises in virus-host interaction.Retrovirology. 2012 Dec 20;9:112. doi: 10.1186/1742-4690-9-112. Retrovirology. 2012. PMID: 23254112 Free PMC article. Review.
-
Life of psi: how full-length HIV-1 RNAs become packaged genomes in the viral particles.Virology. 2014 Apr;454-455:362-70. doi: 10.1016/j.virol.2014.01.019. Epub 2014 Feb 14. Virology. 2014. PMID: 24530126 Free PMC article. Review.
-
From promoting to inhibiting: diverse roles of helicases in HIV-1 Replication.Retrovirology. 2012 Sep 28;9:79. doi: 10.1186/1742-4690-9-79. Retrovirology. 2012. PMID: 23020886 Free PMC article. Review.
References
-
- McLaren M, Marsh K, Cochrane A. Modulating HIV-1 RNA processing and utilization. Front Biosci. 2008;13:5693–5707. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

