Identification of the cluster control region for the protocadherin-beta genes located beyond the protocadherin-gamma cluster
- PMID: 21771796
- PMCID: PMC3173131
- DOI: 10.1074/jbc.M111.245605
Identification of the cluster control region for the protocadherin-beta genes located beyond the protocadherin-gamma cluster
Abstract
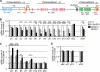
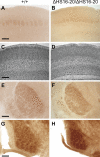
The clustered protocadherins (Pcdhs), Pcdh-α, -β, and -γ, are transmembrane proteins constituting a subgroup of the cadherin superfamily. Each Pcdh cluster is arranged in tandem on the same chromosome. Each of the three Pcdh clusters shows stochastic and combinatorial expression in individual neurons, thus generating a hugely diverse set of possible cell surface molecules. Therefore, the clustered Pcdhs are candidates for determining neuronal molecular diversity. Here, we showed that the targeted deletion of DNase I hypersensitive (HS) site HS5-1, previously identified as a Pcdh-α regulatory element in vitro, affects especially the expression of specific Pcdh-α isoforms in vivo. We also identified a Pcdh-β cluster control region (CCR) containing six HS sites (HS16, 17, 17', 18, 19, and 20) downstream of the Pcdh-γ cluster. This CCR comprehensively activates the expression of the Pcdh-β gene cluster in cis, and its deletion dramatically decreases their expression levels. Deleting the CCR nonuniformly down-regulates some Pcdh-γ isoforms and does not affect Pcdh-α expression. Thus, the CCR effect extends beyond the 320-kb region containing the Pcdh-γ cluster to activate the upstream Pcdh-β genes. Thus, we concluded that the CCR is a highly specific regulatory unit for Pcdh-β expression on the clustered Pcdh genomic locus. These findings suggest that each Pcdh cluster is controlled by distinct regulatory elements that activate their expression and that the stochastic gene regulation of the clustered Pcdhs is controlled by the complex chromatin architecture of the clustered Pcdh locus.
Figures


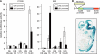



References
-
- Yagi T. (2008) Dev. Growth Differ 50, S131–140 - PubMed
-
- Zipursky S. L., Sanes J. R. (2010) Cell 143, 343–353 - PubMed
-
- Kohmura N., Senzaki K., Hamada S., Kai N., Yasuda R., Watanabe M., Ishii H., Yasuda M., Mishina M., Yagi T. (1998) Neuron 20, 1137–1151 - PubMed
-
- Wu Q., Maniatis T. (1999) Cell 97, 779–790 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

