Ascl1 expression defines a subpopulation of lineage-restricted progenitors in the mammalian retina
- PMID: 21771810
- PMCID: PMC3143566
- DOI: 10.1242/dev.064006
Ascl1 expression defines a subpopulation of lineage-restricted progenitors in the mammalian retina
Abstract
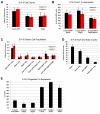
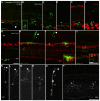
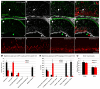
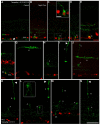
The mechanisms of cell fate diversification in the retina are not fully understood. The seven principal cell types of the neural retina derive from a population of multipotent progenitors during development. These progenitors give rise to multiple cell types concurrently, suggesting that progenitors are a heterogeneous population. It is thought that differences in progenitor gene expression are responsible for differences in progenitor competence (i.e. potential) and, subsequently, fate diversification. To elucidate further the mechanisms of fate diversification, we assayed the expression of three transcription factors made by retinal progenitors: Ascl1 (Mash1), Ngn2 (Neurog2) and Olig2. We observed that progenitors were heterogeneous, expressing every possible combination of these transcription factors. To determine whether this progenitor heterogeneity correlated with different cell fate outcomes, we conducted Ascl1- and Ngn2-inducible expression fate mapping using the CreER™/LoxP system. We found that these two factors gave rise to markedly different distributions of cells. The Ngn2 lineage comprised all cell types, but retinal ganglion cells (RGCs) were exceedingly rare in the Ascl1 lineage. We next determined whether Ascl1 prevented RGC development. Ascl1-null mice had normal numbers of RGCs and, interestingly, we observed that a subset of Ascl1+ cells could give rise to cells expressing Math5 (Atoh7), a transcription factor required for RGC competence. Our results link progenitor heterogeneity to different fate outcomes. We show that Ascl1 expression defines a competence-restricted progenitor lineage in the retina, providing a new mechanism to explain fate diversification.
Figures



References
-
- Akagi T., Inoue T., Miyoshi G., Bessho Y., Takahashi M., Lee J. E., Guillemot F., Kageyama R. (2004). Requirement of multiple bHLH genes for retinal neuronal subtype specification. J. Biol. Chem. 279, 28492-28498 - PubMed
-
- Alvarez-Rodriguez R., Pons S. (2009). Expression of the proneural gene encoding Mash1 suppresses MYCN mitotic activity. J. Cell Sci. 122, 595-599 - PubMed
-
- Anchan R. M., Reh T. A., Angello J., Balliet A., Walker M. (1991). EGF and TGF-alpha stimulate retinal neuroepithelial cell proliferation in vitro. Neuron 6, 923-936 - PubMed
-
- Bassett E. A., Pontoriero G. F., Feng W., Marquardt T., Fini M. E., Williams T., West-Mays J. A. (2007). Conditional deletion of activating protein 2alpha (AP-2alpha) in the developing retina demonstrates non-cell-autonomous roles for AP-2alpha in optic cup development. Mol. Cell. Biol. 27, 7497-7510 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

