A genomic approach to study anthocyanin synthesis and flower pigmentation in passionflowers
- PMID: 21772993
- PMCID: PMC3137904
- DOI: 10.4061/2011/371517
A genomic approach to study anthocyanin synthesis and flower pigmentation in passionflowers
Abstract
Most of the plant pigments ranging from red to purple colors belong to the anthocyanin group of flavonoids. The flowers of plants belonging to the genus Passiflora (passionflowers) show a wide range of floral adaptations to diverse pollinating agents, including variation in the pigmentation of floral parts ranging from white to red and purple colors. Exploring a database of expressed sequence tags obtained from flower buds of two divergent Passiflora species, we obtained assembled sequences potentially corresponding to 15 different genes of the anthocyanin biosynthesis pathway in these species. The obtained sequences code for putative enzymes are involved in the production of flavonoid precursors, as well as those involved in the formation of particular ("decorated") anthocyanin molecules. We also obtained sequences encoding regulatory factors that control the expression of structural genes and regulate the spatial and temporal accumulation of pigments. The identification of some of the putative Passiflora anthocyanin biosynthesis pathway genes provides novel resources for research on secondary metabolism in passionflowers, especially on the elucidation of the processes involved in floral pigmentation, which will allow future studies on the role of pigmentation in pollinator preferences in a molecular level.
Figures



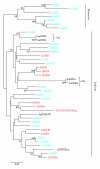
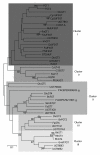


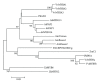

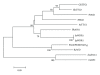
References
-
- Harborne JB. The Flavonoids: Advances in Research Since 1986. 1st edition. New York, NY, USA: Chapman & Hall/CRC; 1994.
-
- Grotewold E. The genetics and biochemistry of floral pigments. Annual Review of Plant Biology. 2006;57:761–780. - PubMed
-
- Rausher MD. Evolutionary transitions in floral color. International Journal of Plant Sciences. 2008;169(1):7–21.
LinkOut - more resources
Full Text Sources

