Tempo and mode of plant RNA virus escape from RNA interference-mediated resistance
- PMID: 21775453
- PMCID: PMC3196453
- DOI: 10.1128/JVI.05326-11
Tempo and mode of plant RNA virus escape from RNA interference-mediated resistance
Abstract
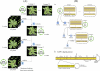
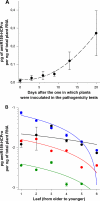
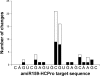
A biotechnological application of artificial microRNAs (amiRs) is the generation of plants that are resistant to virus infection. This resistance has proven to be highly effective and sequence specific. However, before these transgenic plants can be deployed in the field, it is important to evaluate the likelihood of the emergence of resistance-breaking mutants. Two issues are of particular interest: (i) whether such mutants can arise in nontransgenic plants that may act as reservoirs and (ii) whether a suboptimal expression level of the transgene, resulting in subinhibitory concentrations of the amiR, would favor the emergence of escape mutants. To address the first issue, we experimentally evolved independent lineages of Turnip mosaic virus (TuMV) (family Potyviridae) in fully susceptible wild-type Arabidopsis thaliana plants and then simulated the spillover of the evolving virus to fully resistant A. thaliana transgenic plants. To address the second issue, the evolution phase took place with transgenic plants that expressed the amiR at subinhibitory concentrations. Our results show that TuMV populations replicating in susceptible hosts accumulated resistance-breaking alleles that resulted in the overcoming of the resistance of fully resistant plants. The rate at which resistance was broken was 7 times higher for TuMV populations that experienced subinhibitory concentrations of the antiviral amiR. A molecular characterization of escape alleles showed that they all contained at least one nucleotide substitution in the target sequence, generally a transition of the G-to-A and C-to-U types, with many instances of convergent molecular evolution. To better understand the viral population dynamics taking place within each host, as well as to evaluate relevant population genetic parameters, we performed in silico simulations of the experiments. Together, our results contribute to the rational management of amiR-based antiviral resistance in plants.
Figures
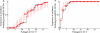
References
-
- Bishop K. N., Holmes R. K., Sheehy A. M., Malim M. H. 2004. APOBEC-mediated editing of viral RNA. Science 305:645. - PubMed
-
- Boucher C. A., et al. 1992. Ordered appearance of zidovudine resistance mutations during treatment of 18 human immunodeficiency virus-positive subjects. J. Infect. Dis. 165:105–110 - PubMed

