An externalized polypeptide partitions between two distinct sites on genome-released poliovirus particles
- PMID: 21775460
- PMCID: PMC3196412
- DOI: 10.1128/JVI.05013-11
An externalized polypeptide partitions between two distinct sites on genome-released poliovirus particles
Abstract
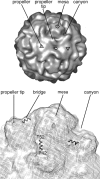
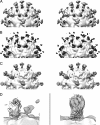
During cell entry, native poliovirus (160S) converts to a cell-entry intermediate (135S) particle, resulting in the externalization of capsid proteins VP4 and the amino terminus of VP1 (residues 1 to 53). Externalization of these entities is followed by release of the RNA genome (uncoating), leaving an empty (80S) particle. The antigen-binding fragment (Fab) of a monospecific peptide 1 (P1) antibody, which was raised against a peptide corresponding to amino-terminal residues 24 to 40 of VP1, was utilized to track the location of the amino terminus of VP1 in the 135S and 80S states of poliovirus particles via cryogenic electron microscopy (cryo-EM) and three-dimensional image reconstruction. On 135S, P1 Fabs bind to a prominent feature on the external surface known as the "propeller tip." In contrast, our initial 80S-P1 reconstruction showed P1 Fabs also binding to a second site, at least 50 Å distant, at the icosahedral 2-fold axes. Further analysis showed that the overall population of 80S-P1 particles consisted of three kinds of capsids: those with P1 Fabs bound only at the propeller tips, P1 Fabs bound only at the 2-fold axes, or P1 Fabs simultaneously bound at both positions. Our results indicate that, in 80S particles, a significant fraction of VP1 can deviate from icosahedral symmetry. Hence, this portion of VP1 does not change conformation synchronously when switching from the 135S state. These conclusions are compatible with previous observations of multiple conformations of the 80S state and suggest that movement of the amino terminus of VP1 has a role in uncoating. Similar deviations from icosahedral symmetry may be biologically significant during other viral transitions.
Figures


Similar articles
-
Cryo-electron microscopy reconstruction shows poliovirus 135S particles poised for membrane interaction and RNA release.J Virol. 2014 Feb;88(3):1758-70. doi: 10.1128/JVI.01949-13. Epub 2013 Nov 20. J Virol. 2014. PMID: 24257617 Free PMC article.
-
The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes.J Virol. 2005 Jun;79(12):7745-55. doi: 10.1128/JVI.79.12.7745-7755.2005. J Virol. 2005. PMID: 15919927 Free PMC article.
-
Conformational shift of a major poliovirus antigen confirmed by immuno-cryogenic electron microscopy.J Immunol. 2013 Jul 15;191(2):884-91. doi: 10.4049/jimmunol.1202014. Epub 2013 Jun 14. J Immunol. 2013. PMID: 23772035 Free PMC article.
-
Structure of the Fab-labeled "breathing" state of native poliovirus.J Virol. 2012 May;86(10):5959-62. doi: 10.1128/JVI.05990-11. Epub 2012 Mar 7. J Virol. 2012. PMID: 22398295 Free PMC article.
-
Hepatitis A Virus Capsid Structure.Cold Spring Harb Perspect Med. 2019 May 1;9(5):a031807. doi: 10.1101/cshperspect.a031807. Cold Spring Harb Perspect Med. 2019. PMID: 30037986 Free PMC article. Review.
Cited by
-
A 3D framework for understanding enterovirus 71.Nat Struct Mol Biol. 2012 Apr 4;19(4):367-8. doi: 10.1038/nsmb.2276. Nat Struct Mol Biol. 2012. PMID: 22472617
-
Cryo-electron microscopy reconstruction shows poliovirus 135S particles poised for membrane interaction and RNA release.J Virol. 2014 Feb;88(3):1758-70. doi: 10.1128/JVI.01949-13. Epub 2013 Nov 20. J Virol. 2014. PMID: 24257617 Free PMC article.
-
Kinetic and structural analysis of coxsackievirus B3 receptor interactions and formation of the A-particle.J Virol. 2014 May;88(10):5755-65. doi: 10.1128/JVI.00299-14. Epub 2014 Mar 12. J Virol. 2014. PMID: 24623425 Free PMC article.
-
RNA transfer from poliovirus 135S particles across membranes is mediated by long umbilical connectors.J Virol. 2013 Apr;87(7):3903-14. doi: 10.1128/JVI.03209-12. Epub 2013 Jan 30. J Virol. 2013. PMID: 23365424 Free PMC article.
-
Structural analysis of coxsackievirus A7 reveals conformational changes associated with uncoating.J Virol. 2012 Jul;86(13):7207-15. doi: 10.1128/JVI.06425-11. Epub 2012 Apr 18. J Virol. 2012. PMID: 22514349 Free PMC article.
References
-
- Baker T. S., Cheng R. H. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryo-electron microscopy. J. Struct. Biol. 116:120–130 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

