Active and total microbial communities in forest soil are largely different and highly stratified during decomposition
- PMID: 21776033
- PMCID: PMC3260513
- DOI: 10.1038/ismej.2011.95
Active and total microbial communities in forest soil are largely different and highly stratified during decomposition
Abstract
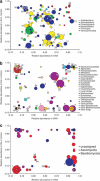
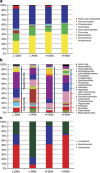
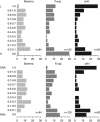
Soils of coniferous forest ecosystems are important for the global carbon cycle, and the identification of active microbial decomposers is essential for understanding organic matter transformation in these ecosystems. By the independent analysis of DNA and RNA, whole communities of bacteria and fungi and its active members were compared in topsoil of a Picea abies forest during a period of organic matter decomposition. Fungi quantitatively dominate the microbial community in the litter horizon, while the organic horizon shows comparable amount of fungal and bacterial biomasses. Active microbial populations obtained by RNA analysis exhibit similar diversity as DNA-derived populations, but significantly differ in the composition of microbial taxa. Several highly active taxa, especially fungal ones, show low abundance or even absence in the DNA pool. Bacteria and especially fungi are often distinctly associated with a particular soil horizon. Fungal communities are less even than bacterial ones and show higher relative abundances of dominant species. While dominant bacterial species are distributed across the studied ecosystem, distribution of dominant fungi is often spatially restricted as they are only recovered at some locations. The sequences of cbhI gene encoding for cellobiohydrolase (exocellulase), an essential enzyme for cellulose decomposition, were compared in soil metagenome and metatranscriptome and assigned to their producers. Litter horizon exhibits higher diversity and higher proportion of expressed sequences than organic horizon. Cellulose decomposition is mediated by highly diverse fungal populations largely distinct between soil horizons. The results indicate that low-abundance species make an important contribution to decomposition processes in soils.
Figures


References
-
- Anderson IC, Parkin PI. Detection of active soil fungi by RT-PCR amplification of precursor rRNA molecules. J Microbiol Methods. 2007;68:248–253. - PubMed
-
- Bååth E, Anderson TH. Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol Biochem. 2003;35:955–963.
-
- Bailey VL, Smith JL, Bolton H. Fungal-to-bacterial ratios in soils investigated for enhanced C sequestration. Soil Biol Biochem. 2002;34:997–1007.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

