Kinome expression profiling and prognosis of basal breast cancers
- PMID: 21777462
- PMCID: PMC3156788
- DOI: 10.1186/1476-4598-10-86
Kinome expression profiling and prognosis of basal breast cancers
Abstract
Background: Basal breast cancers (BCs) represent ~15% of BCs. Although overall poor, prognosis is heterogeneous. Identification of good- versus poor-prognosis patients is difficult or impossible using the standard histoclinical features and the recently defined prognostic gene expression signatures (GES). Kinases are often activated or overexpressed in cancers, and constitute targets for successful therapies. We sought to define a prognostic model of basal BCs based on kinome expression profiling.
Methods: DNA microarray-based gene expression and histoclinical data of 2515 early BCs from thirteen datasets were collected. We searched for a kinome-based GES associated with disease-free survival (DFS) in basal BCs of the learning set using a metagene-based approach. The signature was then tested in basal tumors of the independent validation set.
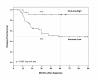
Results: A total of 591 samples were basal. We identified a 28-kinase metagene associated with DFS in the learning set (N = 73). This metagene was associated with immune response and particularly cytotoxic T-cell response. On multivariate analysis, a metagene-based predictor outperformed the classical prognostic factors, both in the learning and the validation (N = 518) sets, independently of the lymphocyte infiltrate. In the validation set, patients whose tumors overexpressed the metagene had a 78% 5-year DFS versus 54% for other patients (p = 1.62E-4, log-rank test).
Conclusions: Based on kinome expression, we identified a predictor that separated basal BCs into two subgroups of different prognosis. Tumors associated with higher activation of cytotoxic tumor-infiltrative lymphocytes harbored a better prognosis. Such classification should help tailor the treatment and develop new therapies based on immune response manipulation.
Figures


Similar articles
-
Sixteen-kinase gene expression identifies luminal breast cancers with poor prognosis.Cancer Res. 2008 Feb 1;68(3):767-76. doi: 10.1158/0008-5472.CAN-07-5516. Cancer Res. 2008. PMID: 18245477
-
A gene expression signature identifies two prognostic subgroups of basal breast cancer.Breast Cancer Res Treat. 2011 Apr;126(2):407-20. doi: 10.1007/s10549-010-0897-9. Epub 2010 May 21. Breast Cancer Res Treat. 2011. PMID: 20490655
-
A Tyrosine Kinase Expression Signature Predicts the Post-Operative Clinical Outcome in Triple Negative Breast Cancers.Cancers (Basel). 2019 Aug 13;11(8):1158. doi: 10.3390/cancers11081158. Cancers (Basel). 2019. PMID: 31412533 Free PMC article.
-
Subtype-dependent prognostic relevance of an interferon-induced pathway metagene in node-negative breast cancer.Mol Oncol. 2014 Oct;8(7):1278-89. doi: 10.1016/j.molonc.2014.04.010. Epub 2014 May 4. Mol Oncol. 2014. PMID: 24853384 Free PMC article.
-
Molecular characterization of breast cancer aggressiveness.Int J Biol Markers. 2003 Jan-Mar;18(1):36-9. doi: 10.1177/172460080301800106. Int J Biol Markers. 2003. PMID: 12699061 Review. No abstract available.
Cited by
-
An in vitro-in vivo model of epithelial mesenchymal transition in triple negative breast cancer.Drug Discov Today Dis Mech. 2012 Summer;9(1-2):e35-e40. doi: 10.1016/j.ddmec.2012.11.002. Drug Discov Today Dis Mech. 2012. PMID: 23585768 Free PMC article.
-
The expression pattern of matrix-producing tumor stroma is of prognostic importance in breast cancer.BMC Cancer. 2016 Nov 4;16(1):841. doi: 10.1186/s12885-016-2864-2. BMC Cancer. 2016. PMID: 27809802 Free PMC article.
-
PD-1/PD-L1 Targeting in Breast Cancer: The First Clinical Evidences Are Emerging. A Literature Review.Cancers (Basel). 2019 Jul 22;11(7):1033. doi: 10.3390/cancers11071033. Cancers (Basel). 2019. PMID: 31336685 Free PMC article. Review.
-
Claudin-low breast cancers: clinical, pathological, molecular and prognostic characterization.Mol Cancer. 2014 Oct 2;13:228. doi: 10.1186/1476-4598-13-228. Mol Cancer. 2014. PMID: 25277734 Free PMC article.
-
A prognostic gene signature for metastasis-free survival of triple negative breast cancer patients.PLoS One. 2013 Dec 11;8(12):e82125. doi: 10.1371/journal.pone.0082125. eCollection 2013. PLoS One. 2013. PMID: 24349199 Free PMC article.
References
-
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lønning PE, Brown PO, Børresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. - DOI - PMC - PubMed
-
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. - DOI - PubMed
-
- van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

