Global functional map of the p23 molecular chaperone reveals an extensive cellular network
- PMID: 21777812
- PMCID: PMC3155841
- DOI: 10.1016/j.molcel.2011.05.029
Global functional map of the p23 molecular chaperone reveals an extensive cellular network
Abstract
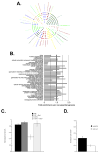
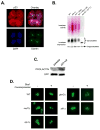
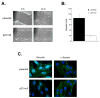
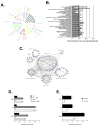
In parallel with evolutionary developments, the Hsp90 molecular chaperone system shifted from a simple prokaryotic factor into an expansive network that includes a variety of cochaperones. We have taken high-throughput genomic and proteomic approaches to better understand the abundant yeast p23 cochaperone Sba1. Our work revealed an unexpected p23 network that displayed considerable independence from known Hsp90 clients. Additionally, our data uncovered a broad nuclear role for p23, contrasting with the historical dogma of restricted cytosolic activities for molecular chaperones. Validation studies demonstrated that yeast p23 was required for proper Golgi function and ribosome biogenesis, and was necessary for efficient DNA repair from a wide range of mutagens. Notably, mammalian p23 had conserved roles in these pathways as well as being necessary for proper cell mobility. Taken together, our work demonstrates that the p23 chaperone serves a broad physiological network and functions both in conjunction with and sovereign to Hsp90.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


Comment in
-
A network of its own: the unique interactome of the Hsp90 cochaperone, Sba1/p23.Mol Cell. 2011 Jul 22;43(2):159-60. doi: 10.1016/j.molcel.2011.07.005. Mol Cell. 2011. PMID: 21777805 Free PMC article.
Similar articles
-
p23/Sba1p protects against Hsp90 inhibitors independently of its intrinsic chaperone activity.Mol Cell Biol. 2008 May;28(10):3446-56. doi: 10.1128/MCB.02246-07. Epub 2008 Mar 24. Mol Cell Biol. 2008. PMID: 18362168 Free PMC article.
-
Hsp90 and p23 Molecular Chaperones Control Chromatin Architecture by Maintaining the Functional Pool of the RSC Chromatin Remodeler.Mol Cell. 2016 Dec 1;64(5):888-899. doi: 10.1016/j.molcel.2016.09.040. Epub 2016 Nov 3. Mol Cell. 2016. PMID: 27818141 Free PMC article.
-
SBA1 encodes a yeast hsp90 cochaperone that is homologous to vertebrate p23 proteins.Mol Cell Biol. 1998 Jul;18(7):3727-34. doi: 10.1128/MCB.18.7.3727. Mol Cell Biol. 1998. PMID: 9632755 Free PMC article.
-
Cdc37 regulation of the kinome: when to hold 'em and when to fold 'em.Sci STKE. 2007 May 8;2007(385):pe22. doi: 10.1126/stke.3852007pe22. Sci STKE. 2007. PMID: 17488976 Review.
-
Expanding the cellular molecular chaperone network through the ubiquitous cochaperones.Biochim Biophys Acta. 2012 Mar;1823(3):668-73. doi: 10.1016/j.bbamcr.2011.08.011. Epub 2011 Aug 24. Biochim Biophys Acta. 2012. PMID: 21889547 Review.
Cited by
-
Gedunin inactivates the co-chaperone p23 protein causing cancer cell death by apoptosis.J Biol Chem. 2013 Mar 8;288(10):7313-25. doi: 10.1074/jbc.M112.427328. Epub 2013 Jan 25. J Biol Chem. 2013. PMID: 23355466 Free PMC article.
-
The Aryl-Hydrocarbon Receptor Protein Interaction Network (AHR-PIN) as Identified by Tandem Affinity Purification (TAP) and Mass Spectrometry.J Toxicol. 2013;2013:279829. doi: 10.1155/2013/279829. Epub 2013 Dec 5. J Toxicol. 2013. PMID: 24454361 Free PMC article.
-
A methylated lysine is a switch point for conformational communication in the chaperone Hsp90.Nat Commun. 2020 Mar 5;11(1):1219. doi: 10.1038/s41467-020-15048-8. Nat Commun. 2020. PMID: 32139682 Free PMC article.
-
Importance of cycle timing for the function of the molecular chaperone Hsp90.Nat Struct Mol Biol. 2016 Nov;23(11):1020-1028. doi: 10.1038/nsmb.3305. Epub 2016 Oct 10. Nat Struct Mol Biol. 2016. PMID: 27723736 Free PMC article.
-
Phosphorylation of p23-1 cochaperone by protein kinase CK2 affects root development in Arabidopsis.Sci Rep. 2019 Jul 8;9(1):9846. doi: 10.1038/s41598-019-46327-0. Sci Rep. 2019. PMID: 31285503 Free PMC article.
References
-
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. - PubMed
-
- Bartkeviciūte D, Sasnauskas K. Disruption of the MNN10 gene enhances protein secretion in Kluyveromyces lactis and Saccharomyces cerevisiae. FEMS Yeast Res. 2004;4:833–840. - PubMed
-
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. - PubMed
-
- DeZwaan DC, Freeman BC. Hsp90: The Rosetta Stone of cellular protein dynamics? Cell Cycle. 2008;7:1006–1012. - PubMed
-
- Ellis J. Proteins as molecular chaperones. Nature. 1987;328:378–379. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

