Pathway and mechanism of drug binding to G-protein-coupled receptors
- PMID: 21778406
- PMCID: PMC3156183
- DOI: 10.1073/pnas.1104614108
Pathway and mechanism of drug binding to G-protein-coupled receptors
Abstract
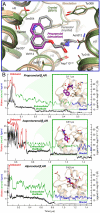
How drugs bind to their receptors--from initial association, through drug entry into the binding pocket, to adoption of the final bound conformation, or "pose"--has remained unknown, even for G-protein-coupled receptor modulators, which constitute one-third of all marketed drugs. We captured this pharmaceutically critical process in atomic detail using the first unbiased molecular dynamics simulations in which drug molecules spontaneously associate with G-protein-coupled receptors to achieve final poses matching those determined crystallographically. We found that several beta blockers and a beta agonist all traverse the same well-defined, dominant pathway as they bind to the β(1)- and β(2)-adrenergic receptors, initially making contact with a vestibule on each receptor's extracellular surface. Surprisingly, association with this vestibule, at a distance of 15 Å from the binding pocket, often presents the largest energetic barrier to binding, despite the fact that subsequent entry into the binding pocket requires the receptor to deform and the drug to squeeze through a narrow passage. The early barrier appears to reflect the substantial dehydration that takes place as the drug associates with the vestibule. Our atomic-level description of the binding process suggests opportunities for allosteric modulation and provides a structural foundation for future optimization of drug-receptor binding and unbinding rates.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Weiland GA, Minneman KP, Molinoff PB. Fundamental difference between the molecular interactions of agonists and antagonists with the β-adrenergic receptor. Nature. 1979;281:114–117. - PubMed
-
- Ward WH, Holdgate GA. Isothermal titration calorimetry in drug discovery. Prog Med Chem. 2001;38:309–376. - PubMed
-
- Swinney DC. Applications of binding kinetics to drug discovery: Translation of binding mechanisms to clinically differentiated therapeutic responses. Pharmaceut Med. 2008;22:23–34.
-
- Baldwin AJ, Kay LE. NMR spectroscopy brings invisible protein states into focus. Nat Chem Biol. 2009;5:808–814. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

