Tirandamycin biosynthesis is mediated by co-dependent oxidative enzymes
- PMID: 21778983
- PMCID: PMC3154026
- DOI: 10.1038/nchem.1087
Tirandamycin biosynthesis is mediated by co-dependent oxidative enzymes
Abstract
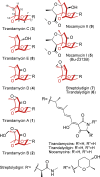
Elucidation of natural product biosynthetic pathways provides important insights into the assembly of potent bioactive molecules, and expands access to unique enzymes able to selectively modify complex substrates. Here, we show full reconstitution, in vitro, of an unusual multi-step oxidative cascade for post-assembly-line tailoring of tirandamycin antibiotics. This pathway involves a remarkably versatile and iterative cytochrome P450 monooxygenase (TamI) and a flavin adenine dinucleotide-dependent oxidase (TamL), which act co-dependently through the repeated exchange of substrates. TamI hydroxylates tirandamycin C (TirC) to generate tirandamycin E (TirE), a previously unidentified tirandamycin intermediate. TirE is subsequently oxidized by TamL, giving rise to the ketone of tirandamycin D (TirD), after which a unique exchange back to TamI enables successive epoxidation and hydroxylation to afford, respectively, the final products tirandamycin A (TirA) and tirandamycin B (TirB). Ligand-free, substrate- and product-bound crystal structures of bicovalently flavinylated TamL oxidase reveal a likely mechanism for the C10 oxidation of TirE.
Figures


References
-
- Royles BJL. Naturally occurring tetramic acids - structure, isolation, and synthesis. Chem. Rev. 1995;95:1981–2001.
-
- Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J. Biol. Chem. 1964;239:2379–2385. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

