Bridging the resolution gap in structural modeling of 3D genome organization
- PMID: 21779160
- PMCID: PMC3136432
- DOI: 10.1371/journal.pcbi.1002125
Bridging the resolution gap in structural modeling of 3D genome organization
Abstract
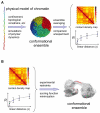
Over the last decade, and especially after the advent of fluorescent in situ hybridization imaging and chromosome conformation capture methods, the availability of experimental data on genome three-dimensional organization has dramatically increased. We now have access to unprecedented details of how genomes organize within the interphase nucleus. Development of new computational approaches to leverage this data has already resulted in the first three-dimensional structures of genomic domains and genomes. Such approaches expand our knowledge of the chromatin folding principles, which has been classically studied using polymer physics and molecular simulations. Our outlook describes computational approaches for integrating experimental data with polymer physics, thereby bridging the resolution gap for structural determination of genomes and genomic domains.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

