Impact of Hfq on global gene expression and virulence in Klebsiella pneumoniae
- PMID: 21779404
- PMCID: PMC3136514
- DOI: 10.1371/journal.pone.0022248
Impact of Hfq on global gene expression and virulence in Klebsiella pneumoniae
Abstract
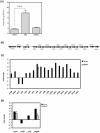
Klebsiella pneumoniae is responsible for a wide range of clinical symptoms. How this bacterium adapts itself to ever-changing host milieu is still a mystery. Recently, small non-coding RNAs (sRNAs) have received considerable attention for their functions in fine-tuning gene expression at a post-transcriptional level to promote bacterial adaptation. Here we demonstrate that Hfq, an RNA-binding protein, which facilitates interactions between sRNAs and their mRNA targets, is critical for K. pneumoniae virulence. A K. pneumoniae mutant lacking hfq (Δhfq) failed to disseminate into extra-intestinal organs and was attenuated on induction of a systemic infection in a mouse model. The absence of Hfq was associated with alteration in composition of envelope proteins, increased production of capsular polysaccharides, and decreased resistance to H(2)O(2), heat shock, and UV irradiation. Microarray-based transcriptome analyses revealed that 897 genes involved in numerous cellular processes were deregulated in the Δhfq strain. Interestingly, Hfq appeared to govern expression of many genes indirectly by affecting sigma factor RpoS and RpoE, since 19.5% (175/897) and 17.3% (155/897) of Hfq-dependent genes belong to the RpoE- and RpoS-regulon, respectively. These results indicate that Hfq regulates global gene expression at multiple levels to modulate the physiological fitness and virulence potential of K. pneumoniae.
Conflict of interest statement
Figures






Similar articles
-
RNA interactome of hypervirulent Klebsiella pneumoniae reveals a small RNA inhibitor of capsular mucoviscosity and virulence.Nat Commun. 2024 Aug 13;15(1):6946. doi: 10.1038/s41467-024-51213-z. Nat Commun. 2024. PMID: 39138169 Free PMC article.
-
The stationary phase sigma factor, RpoS, regulates the production of a carbapenem antibiotic, a bioactive prodigiosin and virulence in the enterobacterial pathogen Serratia sp. ATCC 39006.Microbiology (Reading). 2012 Mar;158(Pt 3):648-658. doi: 10.1099/mic.0.055780-0. Epub 2011 Dec 22. Microbiology (Reading). 2012. PMID: 22194349
-
Contribution of Hfq to photooxidative stress resistance and global regulation in Rhodobacter sphaeroides.Mol Microbiol. 2011 Jun;80(6):1479-95. doi: 10.1111/j.1365-2958.2011.07658.x. Epub 2011 Apr 27. Mol Microbiol. 2011. PMID: 21535243
-
Hfq: a multifaceted RNA chaperone involved in virulence.Future Microbiol. 2016;11(1):137-51. doi: 10.2217/fmb.15.128. Epub 2015 Dec 18. Future Microbiol. 2016. PMID: 26685037 Review.
-
The role of Hfq in bacterial pathogens.Curr Opin Microbiol. 2010 Feb;13(1):24-33. doi: 10.1016/j.mib.2010.01.001. Epub 2010 Jan 14. Curr Opin Microbiol. 2010. PMID: 20080057 Review.
Cited by
-
Identification of Hfq-binding RNAs in Caulobacter crescentus.RNA Biol. 2019 Jun;16(6):719-726. doi: 10.1080/15476286.2019.1593091. Epub 2019 Mar 26. RNA Biol. 2019. PMID: 30870072 Free PMC article.
-
Synthetic Genetic Interactions Reveal a Dense and Cryptic Regulatory Network of Small Noncoding RNAs in Escherichia coli.mBio. 2022 Aug 30;13(4):e0122522. doi: 10.1128/mbio.01225-22. Epub 2022 Aug 3. mBio. 2022. PMID: 35920556 Free PMC article.
-
RNA interactome of hypervirulent Klebsiella pneumoniae reveals a small RNA inhibitor of capsular mucoviscosity and virulence.Nat Commun. 2024 Aug 13;15(1):6946. doi: 10.1038/s41467-024-51213-z. Nat Commun. 2024. PMID: 39138169 Free PMC article.
-
Intrinsic plasmids influence MicF-mediated translational repression of ompF in Yersinia pestis.Front Microbiol. 2015 Aug 21;6:862. doi: 10.3389/fmicb.2015.00862. eCollection 2015. Front Microbiol. 2015. PMID: 26347736 Free PMC article.
-
The RNA chaperone Hfq has a multifaceted role in Edwardsiella ictaluri.Front Cell Infect Microbiol. 2024 Jul 19;14:1394008. doi: 10.3389/fcimb.2024.1394008. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39099884 Free PMC article.
References
-
- Masse E, Salvail H, Desnoyers G, Arguin M. Small RNAs controlling iron metabolism. Curr Opin Microbiol. 2007;10:140–145. - PubMed
-
- Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20:2338–2348. - PubMed
-
- Vanderpool CK. Physiological consequences of small RNA-mediated regulation of glucose-phosphate stress. Curr Opin Microbiol. 2007;10:146–151. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

