The RalGEF-Ral Effector Signaling Network: The Road Less Traveled for Anti-Ras Drug Discovery
- PMID: 21779498
- PMCID: PMC3128631
- DOI: 10.1177/1947601911407329
The RalGEF-Ral Effector Signaling Network: The Road Less Traveled for Anti-Ras Drug Discovery
Abstract
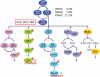
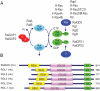
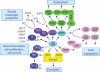
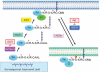
The high frequency of RAS mutations in human cancers (33%) has stimulated intense interest in the development of anti-Ras inhibitors for cancer therapy. Currently, the major focus of these efforts is centered on inhibitors of components involved in Ras downstream effector signaling. In particular, more than 40 inhibitors of the Raf-MEK-ERK mitogen-activated protein kinase cascade and phosphoinositide 3-kinase-AKT-mTOR effector signaling networks are currently under clinical evaluation. However, these efforts are complicated by the fact that Ras can utilize at least 9 additional functionally distinct effectors, with at least 3 additional effectors with validated roles in Ras-mediated oncogenesis. Of these, the guanine nucleotide exchange factors of the Ras-like (Ral) small GTPases (RalGEFs) have emerged as important effectors of mutant Ras in pancreatic, colon, and other cancers. In this review, we summarize the evidence for the importance of this effector pathway in cancer and discuss possible directions for therapeutic inhibition of aberrant Ral activation and signaling.
Keywords: Aurora A; RalBP1/RLIP76; exocyst complex; geranylgeranyltransferase-I inhibitor.
Conflict of interest statement
The author(s) declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
Figures

Similar articles
-
Ral small GTPase signaling and oncogenesis: More than just 15minutes of fame.Biochim Biophys Acta. 2014 Dec;1843(12):2976-2988. doi: 10.1016/j.bbamcr.2014.09.004. Epub 2014 Sep 16. Biochim Biophys Acta. 2014. PMID: 25219551 Free PMC article. Review.
-
Signal pathways which promote invasion and metastasis: critical and distinct contributions of extracellular signal-regulated kinase and Ral-specific guanine exchange factor pathways.Mol Cell Biol. 2001 Sep;21(17):5958-69. doi: 10.1128/MCB.21.17.5958-5969.2001. Mol Cell Biol. 2001. PMID: 11486034 Free PMC article.
-
RAL GTPases: Biology and Potential as Therapeutic Targets in Cancer.Pharmacol Rev. 2018 Jan;70(1):1-11. doi: 10.1124/pr.117.014415. Pharmacol Rev. 2018. PMID: 29196555 Free PMC article. Review.
-
The RalGEF/Ral pathway: evaluating an intervention opportunity for Ras cancers.Enzymes. 2013;34 Pt. B:137-56. doi: 10.1016/B978-0-12-420146-0.00006-8. Epub 2013 Nov 7. Enzymes. 2013. PMID: 25034103 Review.
-
Parallel Rap1>RalGEF>Ral and Ras signals sculpt the C. elegans nervous system.Dev Biol. 2021 Sep;477:37-48. doi: 10.1016/j.ydbio.2021.05.004. Epub 2021 May 13. Dev Biol. 2021. PMID: 33991533 Free PMC article.
Cited by
-
Preclinical-to-clinical Anti-cancer Drug Response Prediction and Biomarker Identification Using TINDL.Genomics Proteomics Bioinformatics. 2023 Jun;21(3):535-550. doi: 10.1016/j.gpb.2023.01.006. Epub 2023 Feb 11. Genomics Proteomics Bioinformatics. 2023. PMID: 36775056 Free PMC article.
-
Conquering oncogenic KRAS and its bypass mechanisms.Theranostics. 2022 Jul 18;12(13):5691-5709. doi: 10.7150/thno.71260. eCollection 2022. Theranostics. 2022. PMID: 35966590 Free PMC article. Review.
-
Localization of RalB signaling at endomembrane compartments and its modulation by autophagy.Sci Rep. 2019 Jun 20;9(1):8910. doi: 10.1038/s41598-019-45443-1. Sci Rep. 2019. PMID: 31222145 Free PMC article.
-
RalGTPases contribute to Schwann cell repair after nerve injury via regulation of process formation.J Cell Biol. 2019 Jul 1;218(7):2370-2387. doi: 10.1083/jcb.201811002. Epub 2019 Jun 14. J Cell Biol. 2019. PMID: 31201266 Free PMC article.
-
RLIP76: A Structural and Functional Triumvirate.Cancers (Basel). 2021 May 4;13(9):2206. doi: 10.3390/cancers13092206. Cancers (Basel). 2021. PMID: 34064388 Free PMC article. Review.
References
-
- Yeh JJ, Der CJ. Targeting signal transduction in pancreatic cancer treatment. Expert Opin Ther Targets. 2007;11:673-94 - PubMed
-
- Buday L, Downward J. Many faces of Ras activation. Biochim Biophys Acta. 2008;1786:178-87 - PubMed
-
- Repasky GA, Chenette EJ, Der CJ. Renewing the conspiracy theory debate: does Raf function alone to mediate Ras oncogenesis? Trends Cell Biol. 2004;14:639-47 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
