Transforming growth factor Beta2 is required for valve remodeling during heart development
- PMID: 21780244
- PMCID: PMC3337781
- DOI: 10.1002/dvdy.22702
Transforming growth factor Beta2 is required for valve remodeling during heart development
Abstract
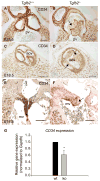
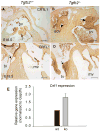
Although the function of transforming growth factor beta2 (TGFβ2) in epithelial mesenchymal transition (EMT) is well studied, its role in valve remodeling remains to be fully explored. Here, we used histological, morphometric, immunohistochemical and molecular approaches and showed that significant dysregulation of major extracellular matrix (ECM) components contributed to valve remodeling defects in Tgfb2(-/-) embryos. The data indicated that cushion mesenchymal cell differentiation was impaired in Tgfb2(-/-) embryos. Hyaluronan and cartilage link protein-1 (CRTL1) were increased in hyperplastic valves of Tgfb2(-/-) embryos, indicating increased expansion and diversification of cushion mesenchyme into the cartilage cell lineage during heart development. Finally, Western blot and immunohistochemistry analyses indicate that the activation of SMAD2/3 was decreased in Tgfb2(-/-) embryos during valve remodeling. Collectively, the data indicate that TGFβ2 promotes valve remodeling and differentiation by inducing matrix organization and suppressing cushion mesenchyme differentiation into cartilage cell lineage during heart development.
Copyright © 2011 Wiley-Liss, Inc.
Figures


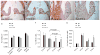

References
-
- Abdelwahid E, Pelliniemi LJ, Jokinen E. Cell death and differentiation in the development of the endocardial cushion of the embryonic heart. Microsc Res Tech. 2002;58:395–403. - PubMed
-
- Amara FM, Entwistle J, Kuschak TI, Turley EA, Wright JA. Transforming growth factor-beta1 stimulates multiple protein interactions at a unique cis-element in the 3′-untranslated region of the hyaluronan receptor RHAMM mRNA. J Biol Chem. 1996;271:15279–15284. - PubMed
-
- Arthur HM, Bamforth SD. TGFbeta signaling and congenital heart disease: Insights from mouse studies. Birth Defects Res A Clin Mol Teratol. 2011:10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

