Biology and physics of von Willebrand factor concatamers
- PMID: 21781248
- PMCID: PMC4117350
- DOI: 10.1111/j.1538-7836.2011.04320.x
Biology and physics of von Willebrand factor concatamers
Abstract
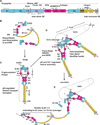
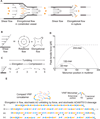
Structural specialisations enable von Willebrand factor (VWF) to assemble during biosynthesis into helical tubules in Weibel-Palade bodies (WPB). Specialisations include a pH-regulated dimeric bouquet formed by the C-terminal half of VWF and helical assembly guided by the N-terminal half that templates inter-dimer disulphide bridges. Orderly assembly and storage of ultra-long concatamers in helical tubules, without crosslinking of neighboring tubules, enables unfurling during secretion without entanglement. Length regulation occurs post-secretion, by hydrodynamic force-regulated unfolding of the VWF A2 domain, and its cleavage by the plasma protease ADAMTS13 (a disintegrin and metalloprotease with a thrombospondin type 1 motif, member 13). VWF is longest at its site of secretion, where its haemostatic function is most important. Moreover, elongational hydrodynamic forces on VWF are strongest just where needed, when bound to the vessel wall, or in elongational flow in the circulation at sites of vessel rupture or vasoconstriction in haemostasis. Elongational forces regulate haemostasis by activating binding of the A1 domain to platelet GPIbα, and over longer time periods, regulate VWF length by unfolding of the A2 domain for cleavage by ADAMTS13. Recent structures of A2 and single molecule measurements of A2 unfolding and cleavage by ADAMTS13 illuminate the mechanisms of VWF length regulation. Single molecule studies on the A1-GPIb receptor-ligand bond demonstrate a specialised flex-bond that enhances resistance to the strong hydrodynamic forces experienced at sites of haemorrhage.
© 2011 International Society on Thrombosis and Haemostasis.
Figures

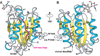
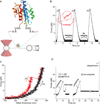
References
-
- Sadler JE. New concepts in von Willebrand disease. Annu Rev Med. 2005;56:173–191. - PubMed
-
- Wagner DD. Cell biology of von Willebrand factor. Annu Rev Cell Biol. 1990;6:217–246. - PubMed
-
- Sadler JE. Biochemistry and genetics of von Willebrand factor. Annu Rev Biochem. 1998;67:395–424. - PubMed
-
- Ruggeri ZM, Mendolicchio GL. Adhesion mechanisms in platelet function. Circ Res. 2007;100:1673–1685. - PubMed
-
- Zenner HL, Collinson LM, Michaux G, Cutler DF. High-pressure freezing provides insights into Weibel-Palade body biogenesis. J Cell Sci. 2007;120:2117–2125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

