Integrating factor analysis and a transgenic mouse model to reveal a peripheral blood predictor of breast tumors
- PMID: 21781289
- PMCID: PMC3178481
- DOI: 10.1186/1755-8794-4-61
Integrating factor analysis and a transgenic mouse model to reveal a peripheral blood predictor of breast tumors
Abstract
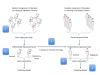
Background: Transgenic mouse tumor models have the advantage of facilitating controlled in vivo oncogenic perturbations in a common genetic background. This provides an idealized context for generating transcriptome-based diagnostic models while minimizing the inherent noisiness of high-throughput technologies. However, the question remains whether models developed in such a setting are suitable prototypes for useful human diagnostics. We show that latent factor modeling of the peripheral blood transcriptome in a mouse model of breast cancer provides the basis for using computational methods to link a mouse model to a prototype human diagnostic based on a common underlying biological response to the presence of a tumor.
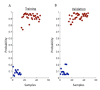
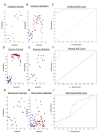
Methods: We used gene expression data from mouse peripheral blood cell (PBC) samples to identify significantly differentially expressed genes using supervised classification and sparse ANOVA. We employed these transcriptome data as the starting point for developing a breast tumor predictor from human peripheral blood mononuclear cells (PBMCs) by using a factor modeling approach.
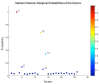
Results: The predictor distinguished breast cancer patients from healthy individuals in a cohort of patients independent from that used to build the factors and train the model with 89% sensitivity, 100% specificity and an area under the curve (AUC) of 0.97 using Youden's J-statistic to objectively select the model's classification threshold. Both permutation testing of the model and evaluating the model strategy by swapping the training and validation sets highlight its stability.
Conclusions: We describe a human breast tumor predictor based on the gene expression of mouse PBCs. This strategy overcomes many of the limitations of earlier studies by using the model system to reduce noise and identify transcripts associated with the presence of a breast tumor over other potentially confounding factors. Our results serve as a proof-of-concept for using an animal model to develop a blood-based diagnostic, and it establishes an experimental framework for identifying predictors of solid tumors, not only in the context of breast cancer, but also in other types of cancer.
Figures
References
-
- Showe MK, Vachani A, Kossenkov AV, Yousef M, Nichols C, Nikonova EV, Chang C, Kucharczuk J, Tran B, Wakeam E. et al. Gene expression profiles in peripheral blood mononuclear cells can distinguish patients with non-small cell lung cancer from patients with nonmalignant lung disease. Cancer Res. 2009;69:9202–9210. doi: 10.1158/0008-5472.CAN-09-1378. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

