PromBase: a web resource for various genomic features and predicted promoters in prokaryotic genomes
- PMID: 21781326
- PMCID: PMC3160392
- DOI: 10.1186/1756-0500-4-257
PromBase: a web resource for various genomic features and predicted promoters in prokaryotic genomes
Abstract
Background: As more and more genomes are being sequenced, an overview of their genomic features and annotation of their functional elements, which control the expression of each gene or transcription unit of the genome, is a fundamental challenge in genomics and bioinformatics.
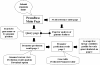
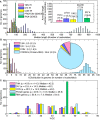
Findings: Relative stability of DNA sequence has been used to predict promoter regions in 913 microbial genomic sequences with GC-content ranging from 16.6% to 74.9%. Irrespective of the genome GC-content the relative stability based promoter prediction method has already been proven to be robust in terms of recall and precision. The predicted promoter regions for the 913 microbial genomes have been accumulated in a database called PromBase. Promoter search can be carried out in PromBase either by specifying the gene name or the genomic position. Each predicted promoter region has been assigned to a reliability class (low, medium, high, very high and highest) based on the difference between its average free energy and the downstream region. The recall and precision values for each class are shown graphically in PromBase. In addition, PromBase provides detailed information about base composition, CDS and CG/TA skews for each genome and various DNA sequence dependent structural properties (average free energy, curvature and bendability) in the vicinity of all annotated translation start sites (TLS).
Conclusion: PromBase is a database, which contains predicted promoter regions and detailed analysis of various genomic features for 913 microbial genomes. PromBase can serve as a valuable resource for comparative genomics study and help the experimentalist to rapidly access detailed information on various genomic features and putative promoter regions in any given genome. This database is freely accessible for academic and non- academic users via the worldwide web http://nucleix.mbu.iisc.ernet.in/prombase/.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

