Resurrection of an ancestral 5S rRNA
- PMID: 21781330
- PMCID: PMC3161009
- DOI: 10.1186/1471-2148-11-218
Resurrection of an ancestral 5S rRNA
Abstract
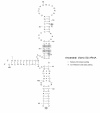
Background: In addition to providing phylogenetic relationships, tree making procedures such as parsimony and maximum likelihood can make specific predictions of actual historical sequences. Resurrection of such sequences can be used to understand early events in evolution. In the case of RNA, the nature of parsimony is such that when applied to multiple RNA sequences it typically predicts ancestral sequences that satisfy the base pairing constraints associated with secondary structure. The case for such sequences being actual ancestors is greatly improved, if they can be shown to be biologically functional.
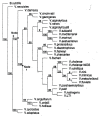
Results: A unique common ancestral sequence of 28 Vibrio 5S ribosomal RNA sequences predicted by parsimony was resurrected and found to be functional in the context of the E. coli cellular environment. The functionality of various point variants and intermediates that were constructed as part of the resurrection were examined in detail. When separately introduced the changes at single stranded positions and individual double variants at base-paired positions were also viable. An additional double variant was examined at a different base-paired position and it was also valid.
Conclusions: The results show that at least in the case of the 5S rRNAs considered here, ancestors predicted by parsimony are likely to be realistic when the prediction is not overly influenced by single outliers. It is especially noteworthy that the phenotype of the predicted ancestors could be anticipated as a cumulative consequence of the phenotypes of the individual variants that comprised them. Thus, point mutation data is potentially useful in evaluating the reasonableness of ancestral sequences predicted by parsimony or other methods. The results also suggest that in the absence of significant tertiary structure constraints double variants that preserve pairing in stem regions will typically be accepted. Overall, the results suggest that it will be feasible to resurrect additional meaningful 5S rRNA ancestors as well as ancestral sequences of many different types of RNA.
Figures
Similar articles
-
Equally parsimonious pathways through an RNA sequence space are not equally likely.J Mol Evol. 1997 Sep;45(3):278-84. doi: 10.1007/pl00006231. J Mol Evol. 1997. PMID: 9302322
-
Common 5S rRNA variants are likely to be accepted in many sequence contexts.J Mol Evol. 2003 Jan;56(1):69-76. doi: 10.1007/s00239-002-2381-6. J Mol Evol. 2003. PMID: 12569424
-
The nucleotide sequence of 5 S ribosomal RNA from Vibrio marinus.Microbiol Sci. 1984 Dec;1(9):229-31. Microbiol Sci. 1984. PMID: 6086080 Review.
-
Defining 5S rRNA structure space: point mutation data can be used to predict the phenotype of multichange variants.Mol Biol Evol. 2011 Sep;28(9):2629-36. doi: 10.1093/molbev/msr090. Epub 2011 Apr 5. Mol Biol Evol. 2011. PMID: 21470967
-
Intermolecular base-paired interaction between complementary sequences present near the 3' ends of 5S rRNA and 18S (16S) rRNA might be involved in the reversible association of ribosomal subunits.Nucleic Acids Res. 1979 Dec 11;7(7):1913-29. doi: 10.1093/nar/7.7.1913. Nucleic Acids Res. 1979. PMID: 94160 Free PMC article. Review.
References
-
- Liberles DA, (editor) Ancestral sequence reconstruction. Oxford University Press; 1997. pp. 1–239.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

