Evaluating target silencing by short hairpin RNA mediated by the group I intron in cultured mammalian cells
- PMID: 21781346
- PMCID: PMC3151216
- DOI: 10.1186/1472-6750-11-79
Evaluating target silencing by short hairpin RNA mediated by the group I intron in cultured mammalian cells
Abstract
Background: The group I intron, a ribozyme that catalyzes its own splicing reactions in the absence of proteins in vitro, is a potential target for rational engineering and attracted our interest due to its potential utility in gene repair using trans-splicing. However, the ribozyme activity of a group I intron appears to be facilitated by RNA chaperones in vivo; therefore, the efficiency of self-splicing could be dependent on the structure around the insert site or the length of the sequence to be inserted. To better understand how ribozyme activity could be modulated in cultured mammalian cells, a group I intron was inserted into a short hairpin RNA (shRNA), and silencing of a reporter gene by the shRNA was estimated to reflect self-splicing activity in vivo. In addition, we appended a theophylline-binding aptamer to the ribozyme to investigate any potential effects caused by a trans-effector.
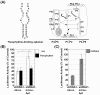
Results: shRNA-expression vectors in which the loop region of the shRNA was interrupted by an intron were constructed to target firefly luciferase mRNA. There was no remarkable toxicity of the shRNA-expression vectors in Cos cells, and the decrease in luciferase activity was measured as an index of the ribozyme splicing activity. In contrast, the expression of the shRNA through intron splicing was completely abolished in 293T cells, although the silencing induced by the shRNA-expressing vector alone was no different from that in the Cos cells. The splicing efficiency of the aptamer-appended intron also had implications for the potential of trans-factors to differentially promote self-splicing among cultured mammalian cells.
Conclusions: Silencing by shRNAs interrupted by a group I intron could be used to monitor self-splicing activity in cultured mammalian cells, and the efficiency of self-splicing appears to be affected by cell-type specific factors, demonstrating the potential effectiveness of a trans-effector.
Figures
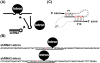



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

