New and old ways to control meiotic recombination
- PMID: 21782271
- PMCID: PMC3177014
- DOI: 10.1016/j.tig.2011.06.007
New and old ways to control meiotic recombination
Abstract
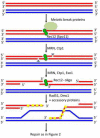
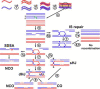
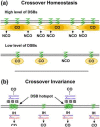
The unique segregation of homologs, rather than sister chromatids, at the first meiotic division requires the formation of crossovers (COs) between homologs by meiotic recombination in most species. Crossovers do not form at random along chromosomes. Rather, their formation is carefully controlled, both at the stage of formation of DNA double-strand breaks (DSBs) that can initiate COs and during the repair of these DSBs. Here, we review control of DSB formation and two recently recognized controls of DSB repair: CO homeostasis and CO invariance. Crossover homeostasis maintains a constant number of COs per cell when the total number of DSBs in a cell is experimentally or stochastically reduced. Crossover invariance maintains a constant CO density (COs per kb of DNA) across much of the genome despite strong DSB hotspots in some intervals. These recently uncovered phenomena show that CO control is even more complex than previously suspected.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
References
-
- Keeney S. Mechanism and control of meiotic recombination initiation. Current Topic Development Biology. 2001;52:1–53. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

